Omicron sublineages, BA.4 and BA.5, were reported from Gauteng in South Africa in April 2022. It's possible that they are fueling a new wave of infections in South Africa, which could eventually erupt worldwide. Both BA.4/5 appear to have evolved from BA.2 and have similar SARS-CoV-2 spike (S) glycoprotein sequences. Additionally, they contain mutations in the S receptor-binding domain (RBD), viz., the R493Q mutation (also found in the SARS-CoV-2 Wuhan-Hu 1 strain), as well as L452R and F486V substitutions.
The substitutions in the BA.4/5 RBDs, L452R, and F486V are of most concern because of their potential to confer immune invasion. Both the mutations are close to the angiotensin-converting enzyme 2 (ACE2) receptor surface, hence can modulate RBD-ACE2 affinity and the neutralizing capacity of natural or vaccine acquired immunity. While the reversion mutation Q493, which also lies within the ACE2, likely reduces the escape from responses to earlier SARS-CoV-2 strains.
About the study
In the present study, researchers reported antigenic characteristics of the Omicron sub-lineages BA.4 and BA.5 (BA.4/5). To this end, the team constructed pseudotyped lentiviruses (pLVs) expressing the S gene of the Omicron sub-lineages BA.1, BA.1.1, BA.2, BA.3, and BA.4/5. They used Wuhan-Hu 1 related strain, Victoria, as the control.
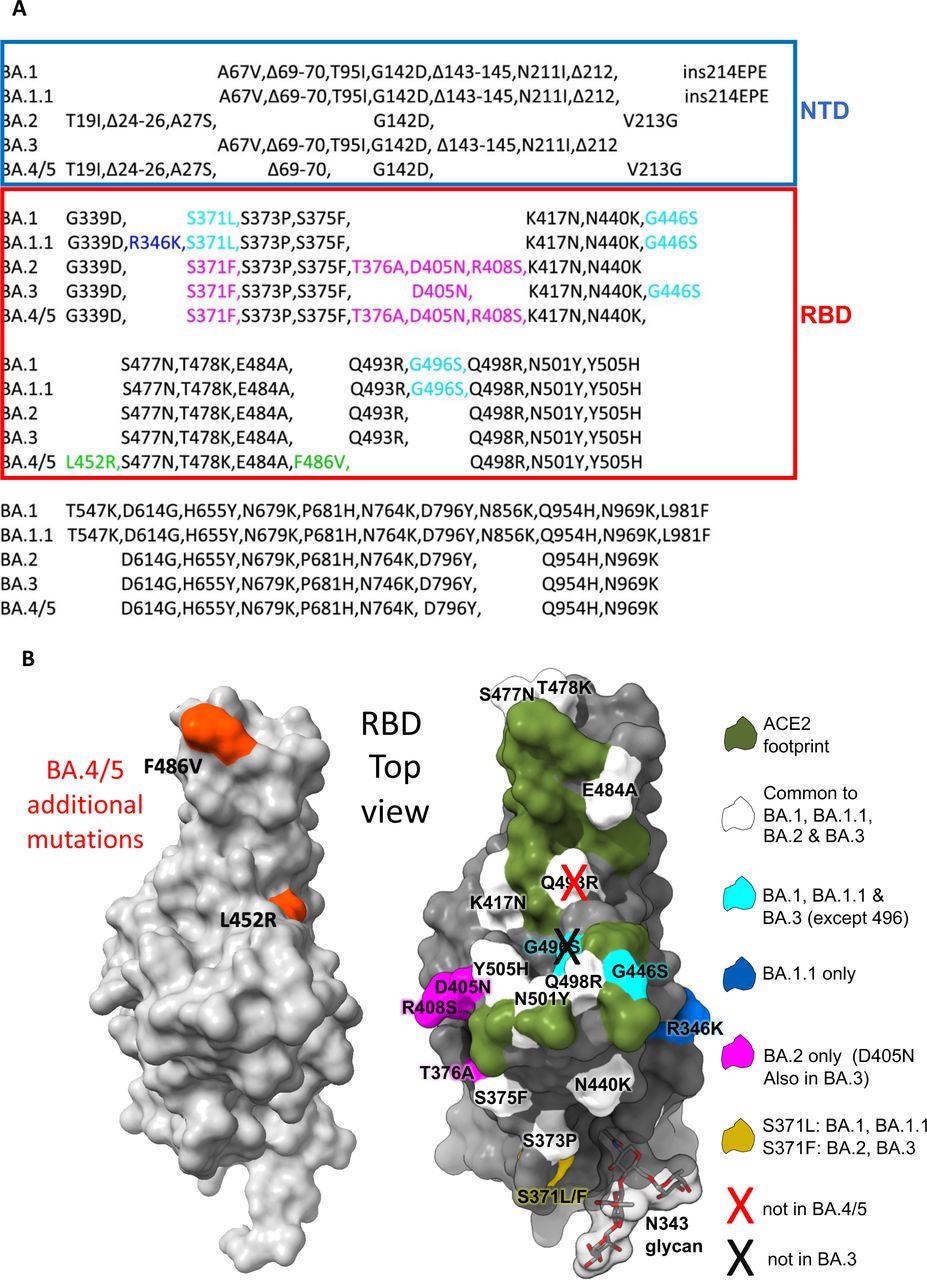
The Omicron sub-lineage compared to BA.4/5. (A) Comparison of S protein mutations of Omicron BA.1, BA.1.1, BA.2, BA.3 and BA.4/5 with NTD and RBD boundaries indicated. (B) Position of RBD mutations (grey surface with the ACE2 footprint in dark green). Mutations common to all Omicron lineages are shown in white (Q493R which is reverted in BA.4/5 is shown with a cross), those common to BA.1 and BA.1.1 in cyan, those unique to BA.1.1 in blue and those unique to BA.2 in magenta. Residue 371 (yellow) is mutated in all Omicron viruses but differs between BA.1 and BA.2. The N343 glycan is shown as sticks with a transparent surface.
Next, the team collected serum samples of coronavirus disease 2019 (COVID-19) vaccinated individuals to perform neutralization assays. Specifically, they obtained 41 and 20 serum samples 28 days following the third dose of the AZD1222 and BNT162b2 vaccine, respectively.
Further, the team collected serum samples from individuals who had suffered breakthrough Omicron infections at two-time points. Accordingly, they collected samples after 14 days from symptom onset and 21 days from symptom onset.
Furthermore, the researchers used surface plasmon resonance (SPR) to test the binding of BA.4/5 and BA.2 RBDs to a panel of therapeutic antibodies. Lastly, the researchers used the neutralization data to place BA.3 and BA.4/5 on an antigenic map.
Study findings
Nutalai et al. have demonstrated that Omicron BA.1 infection following vaccination leads to induction of broadly neutralizing antibodies, with high titers against all the VOCs. In contrast, at the early time point, BA.4/5 titers were reduced 1.9-fold and 1.5-fold compared to BA.1 and BA.2, respectively. At the later time point, BA.4/5 titers were reduced 3.4-fold and two-fold compared to BA.1 and BA.2, respectively. The observations indicate that BA.4/5 escaped both the vaccine-induced and BA.1 infection-induced antibodies more than BA.1 and BA.2 sub-lineages.
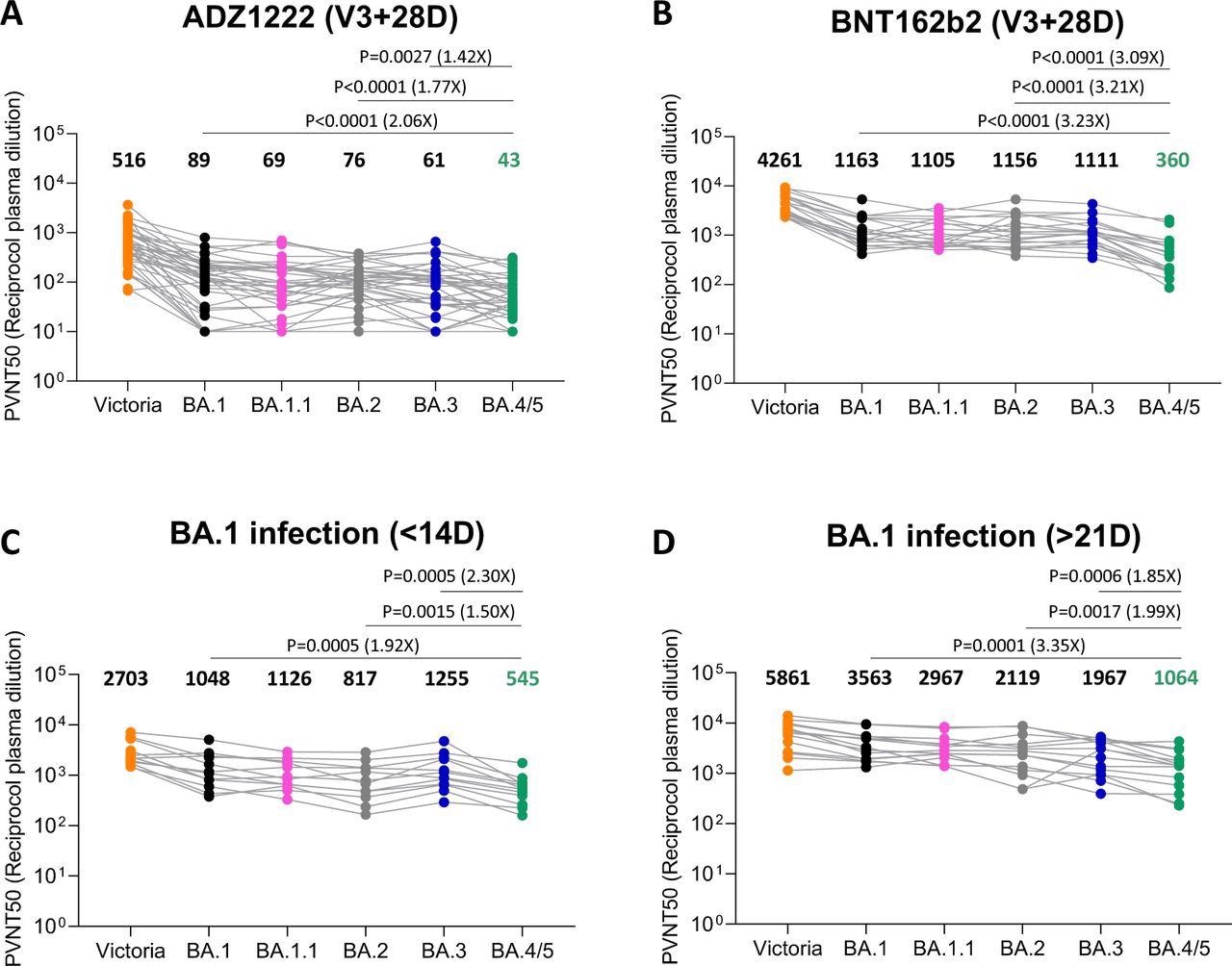
Pseudoviral neutralization assays of BA.4/5 by vaccine and BA.1 immune serum. IC50 values for the indicated viruses using serum obtained from vaccinees 28 days following their third dose of vaccine (A) AstraZeneca AZD AZD1222 (n=41), (B) 4 weeks after the third dose of Pfizer BNT162b2 (n=20). Serum from volunteers suffering breakthrough BA.1 infection volunteer taken (C) early ≤14 (n=12) days from symptom onset (median 13 days) (D) late ≥ 21 days from symptom onset (median 38 days) n=16. Comparison is made with neutralization titres to Victoria an early pandemic strain, BA.1, BA.1..1, BA.2 and BA.3. Geometric mean titres are shown above each column. The Wilcoxon matched-pairs signed rank test was used for the analysis and two-tailed P values were calculated.
In response to the AZD1222 vaccine, neutralization titers for BA.4/5 were reduced 2.1-fold compared to BA.1 and 1.8-fold compared to BA.2. Likewise, the BNT162b2 vaccination reduced neutralization titers by 3.2-fold compared to both BA.1 and BA.2. This data shows that while vaccine efficacy in preventing infection was low, particularly at a later time point as antibody titers wane, it protected progression towards severe disease.
Nutalai et al. also generated a panel of 28 potent human monoclonal antibodies from sera of Omicron cases. Of these, 10 mAbs neutralized BA.4/5 completely, while four other mAbs, Omi-09, 12, 29, and 35, showed a greater than five-fold reduction in the neutralization titer for BA.4/5 compared to BA.2. All the mAbs interacted with the RBD, except Omi-41, which bound the N-terminal domain (NTD). Therefore, it neutralized BA.1, BA.1.1, and BA.3 specifically but not BA.2 or BA.4/5.
SPR analysis confirmed a 1000-fold decrease in the sensitivity of Omi-12 against BA.4/5, an antibody sensitive to the F486V mutation. Likewise, SPR showed a 77-fold enhanced neutralization of BA.4/5 compared to BA.2 for Omi-32, an L452R sensitive antibody. The neutralizing activity of AZD1061 was similar against BA.4/5 and BA.2, while the REG10987, which partially neutralized BA.2, was further weakened against BA.4/5.
The Omicron sub-lineages were clustered together on the antigenic map but remained far from earlier VOCs. Within the Omicron cluster, BA.4/5 was the most distant from the earlier sub-lineages, including BA.1 and BA.2.
Conclusions
The study data demonstrated that Omicron sub-lineages BA.4 and BA.5, compared to BA.1 and BA.2, escaped neutralization from sera of triple vaccinated individuals to a greater extent. Thus, a new or repeat wave of Omicron infection, driven by BA.4/5, is likely in the future. Moreover, the neutralizing activity of many mAbs was either knocked out or drastically impaired against BA.4/5 compared to BA.2.
Overall, the study highlighted that there is plenty of antigenic space for SARS-CoV-2 to explore, and so it may evolve along the Omicron lineage or to a new variant, e.g., Delta to Omicron. SARS-CoV-2 is unlikely to be stopped by COVID-19 vaccination, but next-generation vaccines or vaccines in combination with natural immunity will make people immune to this severe disease.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Further antibody escape by Omicron BA.4 and BA.5 from vaccine and BA.1 serum, Aekkachai Tuekprakhon, Jiandong Huo, Rungtiwa Nutalai, Aiste Dijokaite-Guraliuc, Daming Zhou, Helen M. Ginn, Muneeswaran Sekvaraj, Chang Liu, Alexander J. Mentzer, Piyada Supasa, Helen M. E. Duyvesteyn, Raksha Das, Donal Skelly, Thomas G Ritter, Ali Amini, Sagida Bibi, Sandra Adele, Sile Ann Johnson, Bede Constantinides, Hermione Webster, Nigel Temperton, Paul Klenerman, Eleanor Barnes, Susanna J. Dunachie, Derrick Crook, Andrew J. Pollard, Teresa Lambe, Philip Goulder, OPTIC consortium, ISARIC4C consortium, Elizabeth E Fry, Juthathip Mongkolsapaya, Jingshan Ren, David I. Stuart, Gavin R. Screaton, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.05.21.492554, https://www.biorxiv.org/content/10.1101/2022.05.21.492554v1
- Peer reviewed and published scientific report.
Tuekprakhon, Aekkachai, Rungtiwa Nutalai, Aiste Dijokaite-Guraliuc, Daming Zhou, Helen M. Ginn, Muneeswaran Selvaraj, Chang Liu, et al. 2022. “Antibody Escape of SARS-CoV-2 Omicron BA.4 and BA.5 from Vaccine and BA.1 Serum.” Cell 185 (14): 2422-2433.e13. https://doi.org/10.1016/j.cell.2022.06.005. https://www.cell.com/cell/fulltext/S0092-8674(22)00710-3.