The viral pneumonic illness called COVID-19, caused by the novel coronavirus SARS-CoV-2 has created hotspots of death and severe or critical illness throughout the world. At present, it has claimed over 2.14 million victims, of whom over 143,000 have died. Many of them have been healthcare workers serving in the line of duty.
There is no vaccine, obviously, at present, since the virus was identified only at the end of December 2019. Neither is there any currently known cure. The hope of containing the pandemic rests on the effectiveness of social distancing and lockdown measures, as well as contact tracing and quarantine. These measures are designed to prevent the movement of people who could potentially be carrying the virus with or without symptoms, and who could infect other people through respiratory droplets or other modes of transmission.
While these are meant to ‘flatten the curve’ or reduce the rate at which new cases occur, the only way to actually contain the epidemic would be to find an effective vaccine or therapeutic drug that inhibits viral replication.
Enter the supercomputer center
Researchers at the San Diego Supercomputer Center (SDSC) have recently created a pharmacophore model to help find the potential drugs that could bind the viral protease, the key to the process of viral replication and, therefore, essential for ongoing viral infection. A pharmacophore is an abstract concept, a precise arrangement of common electronic and steric features in the form of atoms, groups or functional groups, that must be present to allow a small molecule to interact specifically with a selected biological target, in order to either stimulate or inhibit the biological response by this binding.
This is one of the chief requirements for a small molecule to be considered a potential drug candidate.
How to find a pharmacophore
Pharmacophore modeling is an application of modern computational chemistry. It enables the investigator to define the core features of different molecules that have the same biological activity. To create a pharmacophore, the scientists look for multiple ligands (molecules with different types of structures) that are bound to the same binding site of the same protein.
For each ligand, the set of 3D low-energy or stable conformational structures that probably includes the bioactive conformation is identified. The ligands are then superimposed or fitted, such that they are aligned by a common feature. The most extensive arrangement that is common to all the ligands is called the common pharmacophore.
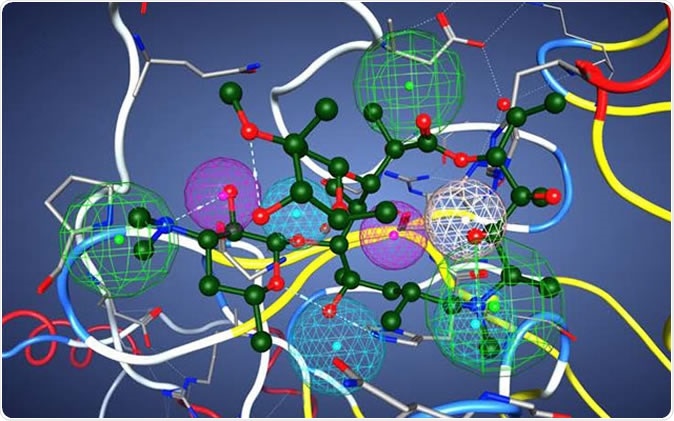
This image illustrates azithromycin in the pharmacophore model of a potential protease inhibitor. The model contains nine functional centers: two donors, two acceptors, one donor/acceptor center, and three hydrophobic centers. Credit: V. Kouznetsova, I. Tsigelny, D. Huang, San Diego Supercomputer Center/UC San Diego
What protease inhibitors do
The investigators explain the principle of the current approach. Likening the host cell to a fortress under siege, scientist Valentina Kouznetsova says multiple lines of defense are available to the cell. For one, the cell surface receptor called ACE2 turns out to be the point of attachment of the virus to the cell. The scientists explain that there could be inhibitors or antibodies which prevent such binding.
Kouznetsova says these are the first line of defense, but the second line of defense is represented by protease inhibitors – drugs that inhibit the key viral enzyme RNA polymerase. Without this enzyme, the virus cannot replicate.
She explains, “To help visualize this, it is as if people on the fortress moor are throwing cobblestones and pouring hot tar on the heads of enemies. The second line of defense is when the enemies arrive, and hand-to-hand combat begins. This latter line is what protease inhibitors do – they don’t allow the virus to produce needed viral proteins for a further attack.”
What did the study do?
The pharmacophore model allows researchers to search a database of many different chemicals, looking for a common set of electronic and steric features with the same orientation relative to each other.
The aim of the current study was to identify which of the FDA-approved drugs are suitable for immediate clinical trials on human patients. This approach has been validated by other human research involving COVID-19, as it cuts the time required for drug approvals by seeking to repurpose already approved drugs.
Professor Roberto Burioni, virologist and author of the new book Virus: The Great Challenge: From Coronavirus to the Plague: How Science Can Save Mankind, says regarding this approach: “To fight an epidemic, you need speed and strategy. The later the reaction, the more likely the defeat.”
In the current scenario, the researchers took the molecular conformations of potential inhibitors and put them through a similarity cluster test, using 3D fingerprints of the molecule. They also allowed several possible binding or conforming molecules to dock to the binding pocket of the protease, and then repeated the docking with any given compound.
Describing the results, researcher Igor Tsigelny says, “This cocktail of inhibitors could help save thousands of lives,” said Tsigelny. “We have now published our findings with Chemrxiv so that we can move on with proper testing that would next allow this concept to be utilized during the current pandemic crisis.”
Journal reference:
Kouznetsova, Valentina; Huang, David; Tsigelny, Igor F. (2020): Potential COVID-19 Protease Inhibitors: Repurposing FDAapproved Drugs. ChemRxiv. Preprint. https://doi.org/10.26434/chemrxiv.12093900.v1