The coronavirus, medically known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is responsible for the global pandemic that has raged for the last five months, sweeping almost the entire populated world, bringing entire economies to a grinding halt, and causing hundreds of thousands of deaths. Originating in Wuhan, China, COVID-19 spread with near-unprecedented speed across the globe, and no cure or vaccine has been discovered yet.
In the face of this bleak picture, a team of researchers from Naples, Italy, has come up with an approach that could help prevent further infections by the SARS-CoV-2.
The Spike Protein
SARS-CoV-2, like other coronaviruses, depends on a glycosylated protein named “Spike” (or Protein S) on its exterior to infiltrate the cells of the human host. Once inside, the virus takes over the host cell machinery to replicate its genome and other essential components. These are assembled within the cell and then emerge from the cell to infect other cells. The cycle continues and may cause the host to fall sick.
The viral particles usually target the epithelial cells in the respiratory tract. Angiotensin-converting enzyme 2 (ACE2) receptors are not the only way a virus can enter a cell, though. Recently, another receptor named CD147 (or Basigin) has been identified as a possible route of entrance. Here too, the virus uses Spike to gain access to the cell. Therefore, Spike is a promising target for antibodies and other potential treatments.
SARS-CoV-2 viruses binding to ACE-2 receptors on a human cell, the initial stage of COVID-19 infection, conceptual 3D illustration Credit: Kateryna Kon / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The ‘Fatal Mimic’ Strategy
The team decided to interrupt the viral entry sequence near its beginning. Spike has two subunits: S1 and S2. The S protein homotrimers are used by the virus to attach to the angiotensin-converting enzyme 2 (ACE2) receptor on the cell membrane, via the receptor-binding domain (RBD) on the S1 subunit.
The current study was focused on creating a molecule that would interact with the S protein. This would act against all known coronaviruses using this receptor, both past, and future.
Using the existing data on the interaction between the SARS-CoV-2 S protein and the ACE2 molecule, the team created a mini-protein, “a miniaturized mimic” of ACE2, which they named Spikeplug. The molecular modeling was based on the cryo EM structure of the SARS-CoV-2 S protein, as well as the crystallization-derived structure of the ACE2 receptor-RBD complex.
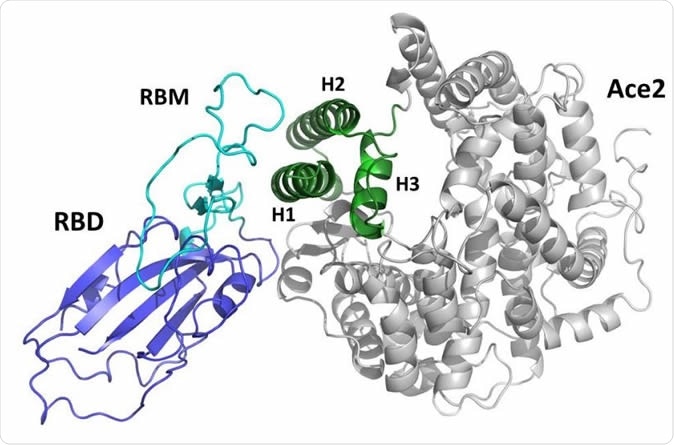
Representation of the complex between the receptor-binding (RBD) domain of SARS-CoV2 Spike protein (blue/cyan) and the human ACE2 receptor (grey/green). The receptor-binding motif (RBM) is drawn in cyan. The green portion of the ACE2 domain including helices H1, H2, and H3 is drawn in green.
They included a number of mutations for greater stability and solubility. For instance, they included an extra helix to stabilize the conformational folding stability, because peptides in solution are not typically folded.
The mini-protein, which they produced in high yield (70 mg pure protein /L of E. coli bacterial culture) using recombination techniques, has a stable alpha-helical conformation and is highly soluble and stable in solution.
Spikeplug embeds all the essential components of ACE2, which are targeted by the S Protein. It was designed to interact and fuse with Spike and therefore prevent it from binding with ACE2 sites on host cells, in effect immobilizing it.
The high-affinity protein can bind to the RBD domain of the glycosylated S protein when present at concentrations of a few nanomoles. Spikeplug is large enough to cover most important interactions with Spike but small enough to bind to all three of its chains.
By effectively preventing the virus from infiltrating a host cell, Spikeplug is a valuable tool in the development of coronavirus treatments.
How is Spikeplug Better Than Other Antivirals?
The team believes that host receptor mimic-based approaches are advantageous over other methods. Firstly, if another coronavirus should emerge and attempt to target ACE2, it will be deterred by the mimic protein.
The second advantage lies in the field of antimicrobial resistance. Most pathogens can mutate relatively quickly to bypass existing treatments. However, since binding to the ACE2 site is a fundamental step in the coronavirus chain of infection, mutating to avoid this step would likely result in a nonviable virus.
This approach has been demonstrated to be effective in the case of the drug Enfuvirtide. This antiretroviral drug works as an entry inhibitor for the HIV virus by inhibiting the action of the gp41 fusion protein. Enfuvirtide can select for resistance mutation: however, such resistant variants are significantly less fit since they must make a significant change in the fusion complex.
Finally, Spikeplug is unlikely to provoke adverse reactions from the body due to its similarity to the body’s native ACE2 protein. The study concludes, “We believe that Spikeplug is a first-in-class promising lead candidate for the development of molecularly targeted therapies against SARS-CoV-2 and other coronaviruses.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Romano, M., et al. (2020). An Engineered Stable Mini-Protein to Plug SARS-Cov-2 Spikes. bioRxiv preprint doi: https://doi.org/10.1101/2020.04.29.067728. https://www.biorxiv.org/content/10.1101/2020.04.29.067728v1
- Peer reviewed and published scientific report.
Squeglia, Flavia, Maria Romano, Luciana Esposito, Giovanni Barra, Pietro Campiglia, Marina Sala, Maria Carmina Scala, Alessia Ruggiero, and Rita Berisio. 2022. “Structure-Based Development of SARS-CoV-2 Spike Interactors.” International Journal of Molecular Sciences 23 (10): 5601. https://doi.org/10.3390/ijms23105601. https://www.mdpi.com/1422-0067/23/10/5601