The frighteningly rapid, almost casual, the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes the pneumonic illness COVID-19 disease, has shaken the nerves of many people. The ease with which this illness spreads and the relatively high fatality rate, especially among the sick and elderly, has raised the public perception of the risks posed by social contacts.
The detection of viral particles in wastewater is not only crucial for virus containment but also the assessment of the spread of the infection – wastewater-based epidemiology. This is because the conventional way of measuring spread, based on symptomatic patients, leaves out the asymptomatic infected individuals who make up a considerable chunk of the infected population.
The current approach was mooted from the observation that the virus lives for a significant period in fecal samples of infected patients. With this in mind, the identification and quantification of viral RNA in wastewater before treatment could accurately indicate the prevalence of infection in the population.
The unknowns in this situation include the time of persistence of coronaviruses, which are enveloped viruses, and, therefore, less robust; and how they spread in water collected and treated by the wastewater network. Some studies have shown recently that the virus could still be not only present but also infectious in treated wastewater.
How was the study done?
The current study took place in northern Italy, which witnessed the earliest and most significant outbreak of infection. As on April 27, 2020, there were 200,000 cases, with an unusually high infection rate that was twice that of the rest of Italy.
The researchers had four aims:
- Detect the presence of the virus and find how effective these plants were at decreasing or clearing the viral load
- Test virus vitality before and after passage through the treatment plants to find its intrinsic persistence
- Compare the strains found in these samples to those obtained from sick patients in the same area and different global locations
- Look for the viral RNA in rivers downstream of the metropolitan area
The researchers looked for the presence of SARS-CoV-2 in raw and treated wastewater samples from 3 wastewater plants. These covered one whole metropolitan area, parts of which had a high population density and were highly urbanized.
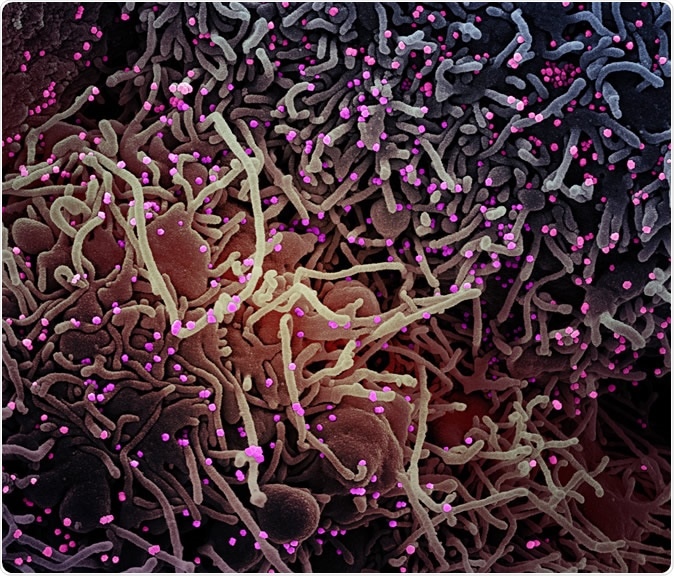
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of a VERO E6 cell (purple) exhibiting elongated cell projections and signs of apoptosis, after infection with SARS-COV-2 virus particles (pink), which were isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The samples were taken from 3 wastewater plants, each of which drain and treat about 11 m3 of sewage from about 2 million people, using secondary and tertiary treatments. They emptied into two rivers.
All samples were taken on April 14 and April 22, 2020, at 1:00 pm. Viral extraction and detection by RT-PCR were carried out, looking for any of six strains of the novel coronavirus, the ORF1ab gene, and the E gene.
Whole-genome sequencing and genome assembly were carried out, followed by phylogenetic analysis.
Virus vitality was done in cell culture.
SARS-CoV-2 in wastewater samples
The raw wastewater samples from all treatment plants showed the presence of the virus on April 14, 2020, but only one sample on April 22, 2020. After treatment, no sample returned positive results. This shows the treatments were effective in reducing viral load almost entirely up to 4 log 10 for many types of virus, and in the case of SARS-CoV-2, by 100-fold reduction from an initial value of 1 x 107 in the raw sample.
The caveat is that the presence of low numbers of viral copies could not be excluded below the sensitivity of the multiplex reaction. In fact, tailoring of the protocols to allow virus concentration from environmental samples and RNA quantification.
If only the inflow concentrations were examined, a concentration of 50-1,500 copies of the nucleocapsid gene/mL was obtained. Since the PCR kit used had a sensitivity level of 1,000 viral copies/mL, this indicates that the wastewater in the metropolitan area was heavily contaminated.
The samples taken eight days later from the inlets of the plants showed that the viral load had possibly decreased since the samples were no longer positive.
How were the wastewater results interpreted?
The daily trends of incidence in the provinces covered in the current study showed that both had similar trends and similar magnitudes. The number of cases began to move up at the end of February 2020, peaked in the second half of March 2020, and slowly fell after that. This data, coming from the national survey service, is limited by the non-standardization of collection efforts, which could affect the ability to link wastewater and clinical, epidemiological results.
The similarity between the study data and the survey data does, however, suggest the feasibility of this approach in estimating infection spread and prevalence. It is worth noting that for this purpose, more sensitive approaches are needed for quantitative PCR by concentrating the viral samples.
Even though there was a high number of RNA copies in the samples, the vitality of the virus in both raw and treated samples was insignificant. Cell cultures showed a lack of cytopathic effects at 48 and 72 hours from inoculation. It is known that enveloped viruses lose infectiveness in wastewater more rapidly than non-enveloped viruses, and especially when there is free enzyme activity or if there are protozoa or metazoan in the water, which prey on these microbes.
Coronaviruses also survive longer at cold temperatures, up to 36 hours at 10°C, but only about 7-13 hours at 23-25°C. In the current study, the arrival of feces at the wastewater plant was not more than 6-8 hours after their passage from the host. The period of collection was also during a mild spell, which prevented the survival of the virus up to the point of reaching the plants.
Overall, it would seem that accidental contact with wastewater in the form of aerosols or droplets does not carry a risk of infection in this study.
SARS-CoV-2 in river samples
The same trend was found in the river samples – both showed the presence of the virus on April 14 but only one sample on April 22. The researchers explain why the virus was present in river water despite being absent in treated wastewater, by the hypothesis that the surface of the river water had a dose of raw wastewater. Since the sampling period coincided with a long and unexpected drought, the concentration of this aliquot was probably exacerbated, they say.
The untreated water could be from household sewage, which did not flow into the main sewage system and thence to the treatment plant, or it could be that urban runoff and household sewage were not adequately separated, leading to combined sewer overflows (CSOs).
CSOs are apparently common in central Europe, where 70% of systems operate this way. In the US, too, 40 million people have similar sewage systems. With a combined sewer system, CSOs are usual during seasons of high rain, to prevent the wastewater treatment plants from being damaged by excessive inflows.
During drought, too, sewage system failures can cause this type of overflow. The correct explanation should be sought using substances like caffeine, which originate in human activities, to trace the origin of the untreated household sewage.
On a positive note, the contamination of the single positive river sample was not an indicator of infectious risk because of the deficient vitality of the virus recovered.
Phylogenetic analysis of SARS-Cov-2 strains
Only one genome from those recovered was sequenced and was closely linked to one isolated in Milan on March 3, 2020. There were two single nucleotide polymorphisms between the strains. This strain is part of the European clade, indicating it comes from the same source.
How was the study important?
The study detected the presence of the virus in wastewater samples and river water samples. As a result, the study concludes, “Wastewater-based epidemiology for SARS CoV2 appears as a promising tool for epidemic trend monitoring to complement current clinical data.”
Moreover, the presence of the virus in the rivers means that the sewerage systems now being used are only partially effective, reflecting a similarity between European and US sewage systems. Nonetheless, the low vitality means that the virus is hardly pathogenic, making the infective risk a negligible one.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources