The use of machine learning helped surmount obstacles posed by the variable immune response in different individuals and identify the underlying gene expression profiles amid all the other noise. They used patient cohorts across many viral pandemics for this purpose, exploiting the resulting pattern of gene expression to study the immune response in COVID-19.
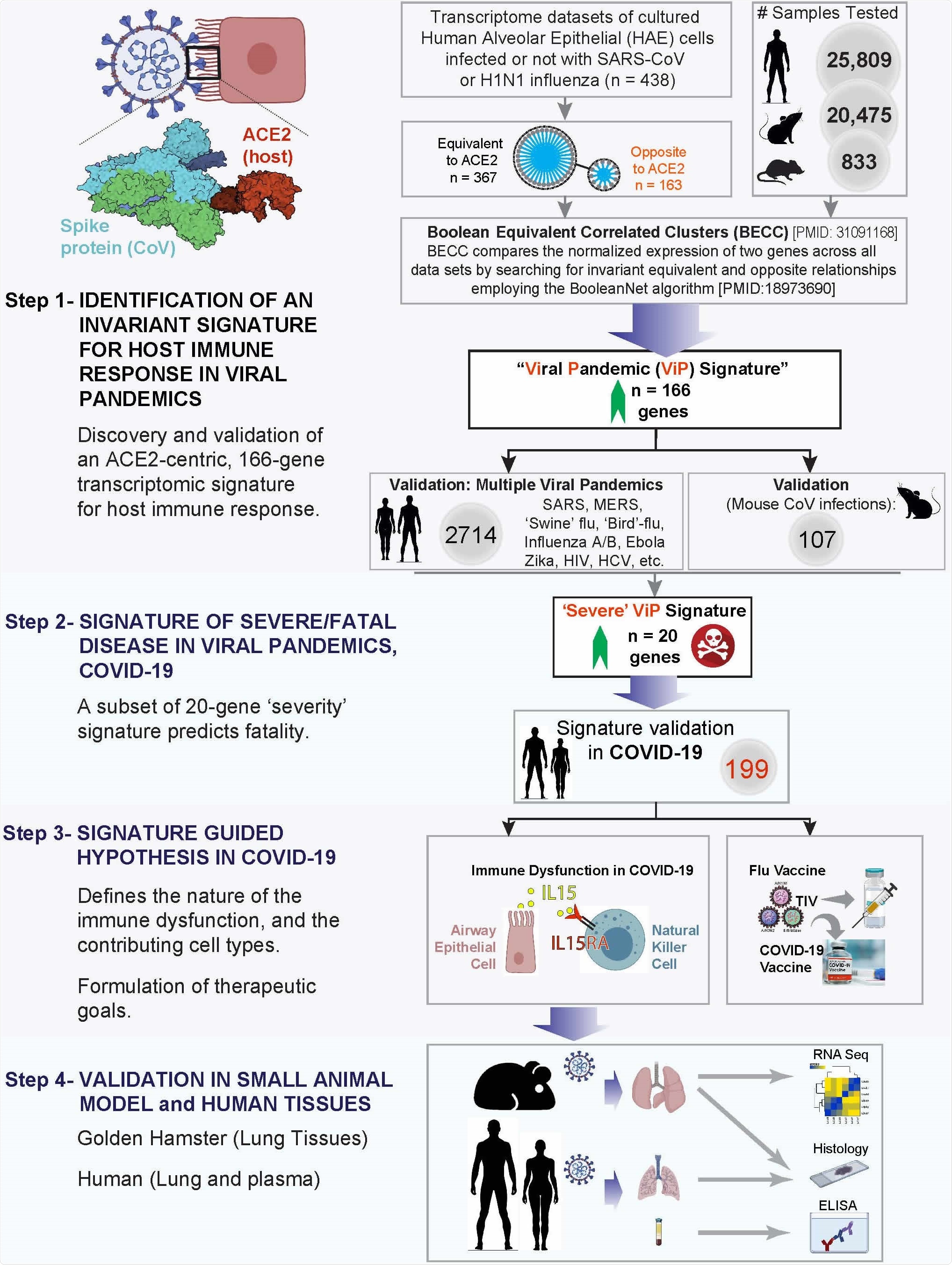
Study design. (From top to bottom) Step 1: A database containing > 45,000 human, mouse and rat gene– expression data was mined to identify and validate an invariant signature for host response to viral pandemic (ViP) infection. ACE2, the portal for SARS-CoV-2 entry/uptake, was used as a ‘seed’ gene and Boolean Equivalent Correlated Clusters (BECC) was used as the computational method to identify gene clusters that share invariant relationships with ACE2. Once defined, these gene clusters (a.k.a., ‘ViP signature’) were subsequently validated across numerous human and murine models of pandemic viral infection. Step 2: A subset of 20 genes from the ViP signature was selected that was strongly associated with severity of viral infection. These genes were validated in other cohorts to establish the ‘Severe’ ViP signature. Both 166- and 20-gene ViP signatures were validated on COVID-19 datasets. Step 3: Cross-validation studies in numerous other datasets helped- (i) define the nature (ii) and source of the cytokine storm in COVID-19, (iii) gain insights into the immunopathology of fatal disease, and (iv) set precise therapeutic goals. Step 4: Findings in step 3 were validated in hamsters and in a cohort of COVID-19 patients.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
ACE2-centric Study Design
In order to find a validated signature for the current pandemic from over 45,000 datasets of gene expression data in humans, mice, and rats, in multiple pandemics, the researchers used the angiotensin-converting enzyme 2 (ACE2) as ‘seed’ gene in the computational approach.
This was for three reasons. Firstly, it is the viral receptor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and its expression reflects innate immune gene expression and tissue susceptibility to the virus. Secondly, it effectively inhibits the renin-angiotensin-aldosterone system (RAAS), a potent pro-inflammatory system. Thirdly, it is disrupted by infections, and this leads to unregulated inflammatory activity.
Informatics Approach
The researchers used a machine learning tool called Boolean Equivalent Correlated Clusters (BECC) in order to find the underlying invariant gene expression relationships in respiratory viral pandemics. They identify the immense advantages of this approach thus:” The BECC algorithm focuses exclusively on “Boolean Equivalent” relationships to identify potentially functionally related gene sets. Once identified, these invariant relationships have been shown to spur new fundamental discoveries with translational potential, and most importantly, offer insights that aid the navigation of uncharted territories where nothing may be known.”
ViP Signature Characteristic of Viral ‘Cytokine Storm’
The researchers found that 166 genes were consistently related to ACE2, with a decline in expression during convalescence. This pattern occurred uniformly across all viral pandemics (not necessarily respiratory virus pandemics), for which reason it was named the ‘Viral Pandemic’ (ViP) Signature.
The majority of the 166 genes were involved in immune responses, including interferon and cytokine signaling within the innate immune system, as well as the adaptive immune system. Thus, they indicated the host immune response to a viral infection per se, as expected, since the cytokine storm in any viral pandemic is known to be due to an exuberant host immune response.
Surprisingly, however, this pattern was detected despite the ACE2-centric approach, though ACE2 is not the viral entry receptor for influenza. It continued unchanged despite filtering through unrelated datasets in vitro and in vivo. Again, the cytokine-receptor pair IL15/IL15RA was found to be invariably associated with ACE2 expression regardless of the dataset.
This agrees with previous findings that IL15 is central in lung injury and determines its severity, while its deletion protects mice from lethal influenza.
The signature was also associated with ACE2-expressing lung epithelium and myeloid cells, indicating a close link between ACE2-mediated viral entry and ACE2-associated host gene activation.
20-Gene Subset Detects Disease Severity
Ranking of the 166 genes for their ability to distinguish severe or critical influenza led to the emergence of a subset of 20 genes that could accurately classify healthy and infected patients as well as mild and severe disease. This subset marks DNA damage, stress-induced senescence, and cell cycle alterations as well as neutrophil degranulation, thus indicating disease severity and death.
The 166-gene ViP signature classified healthy vs. infected patients in all five peripheral blood cells, but the subset was most accurate in neutrophils. The researchers say this indicates that “the cells of the innate immune system are the primary contributors of disease severity.”
COVID-19 Induces ViP Signatures in Lung Epithelium and Immune Cells
Both 166-gene and 20-gene signatures perfectly classified healthy and SARS-CoV-2-infected samples in vitro in all tissues and all lung cell types. The former also distinguished these sets in alveolar epithelium, macrophages, CD4 cells, and NK cells. It was also able to distinguish mild and severe COVID-19 in the first and last cell types.
The 20-gene subset recapitulated these findings best with the same two cell types. This indicates, the authors say, that the signature of disease severity and fatality was prominently expressed in lung epithelium, which is a source of virus-induced IL15, and NK cells, which are the primary IL15 target.
NK cells exposed to virally infected epithelial cells showed both these signatures, thus explaining the findings in COVID-19 patients, and indicating that these could be attributed solely to the effects of IL15.
IL15 Storm Mediates COVID-19 Severity
The obvious association of the two ViP signatures with IL15/IL15RA in the same organ, namely, the lung, suggests the one causes the other. As predicted by these findings, severe COVID-19 is defined most often by exaggerated IL15-dominated cytokine release beginning in the lung, as shown by examining samples from a group of symptomatic COVID-19 patients with varying clinical severity (including autopsy specimens from those who died).
Immunopathogenetic Model of COVID-19
The study shows that the immune system plays a significant role in the pathogenesis of COVID-19. The disease begins with the activation of 166 genes in airway epithelial cells as well as myeloid lineage cells, and other immune cells, leading to IL-15 secretion.
This is supported by earlier studies in which the bronchial cells were shown to express IL15 and IL15RA/B genes constitutively. The synthesis and secretion of IL15 are induced by IFNγ as well. These studies indicate that prolonged and over-intense exposure of NK cells to cytokines leads to their exhaustion and death.
This agrees with recent findings that NK cells are seen to be exhausted and reduced in number in severe COVID-19, as early as 3-6 days from symptom onset.
The authors say, “Fatal COVID-19 is characterized by a paradoxical immune response, i.e., suppression of epithelial and NK cell functions (immunosuppression) in the setting of a cytokine storm (overzealous immune response).”
ViP Profiles Shape Therapy and Indicate Efficacy of Treatment
These researchers had earlier shown that the 166-gene ViP signatures were attenuated during the convalescence period of several respiratory virus pandemics. This finding was demonstrated by the current researchers, who went on to explore their role as an indicator that treatment was causally associated with this response.
To answer this question, they examined earlier interventional studies for HCV, HIV, Zika, and Ebola, involving the use of effective anti-virals. In all these cases, this signature accurately categorized treated vs. non-treated samples, with the former showing attenuation of the ViP signature.
For instance, liver biopsies taken from HCV patients were classified using this signature into those treated with directly acting anti-viral agents (DAAs) and those not treated. Similar results were found with HIV patient samples, treated or not with antiretroviral drugs.
The scientists say, “These findings suggest ‘causality’ between treatment (anti-viral therapies) and response (attenuation of the ViP signature), and imply that attenuation of the 166-gene ViP signature is a desirable therapeutic goal.”
SARS-CoV-2 Virus Is Responsible for The Host Response
The next question was to relate the host immune response to the SARS-CoV-2 virus. They used hamsters treated with anti-SARS-CoV-2 antibody to completely block viral binding to the ACE2 receptor and then challenged with the virus, comparing them with another group pretreated with a control antibody.
They found that firstly, the test group showed neither the 166-gene or the 20-gene ViP signatures so characteristic of the infected lungs. Secondly, the absence of these signatures accompanied the absence of excessive immune cell infiltration or the obliteration of alveolar space. Finally, IL15 and IL15 receptor expression were markedly lower in these tissues.
The researchers feel that this bears out the ACE2-centric approach, showing that when an antibody prevents ACE2-virus binding, the ViP signature and IL 15 cytokine storm are suppressed, while effective treatment reverses both.
Shared Host Response Signature
The researchers point out a “major and unexpected finding,” which is that all viral pandemics have the same host immune response, though the severity, clinical features, etiological virus, and lethality may be entirely different. This may indicate the fundamental importance of ACE2 expression for the host immune response in most viral infections.
Not only does the ViP signature characterize the host immune response to a viral pandemic, but also offers a way to monitor the occurrence and extent of such a response.
The study also showed that the cytokine storm in all pandemics was mediated by uncontrolled IL15 and IL15RA activation. The higher the level of IL15, the more severe the disease was. IL15 levels were significantly higher in men of advanced age, who are known to be a high-risk group for severe COVID-19 worldwide. The lung epithelium and myeloid cells are the chief contributors to the ViP signature and IL15/IL15RA.
Finally, they detected a subset of 20 genes, a signature suggesting severe disease in all viral pandemics.
In their words, “The ViP signatures begin to paint a picture of ‘paradoxical immunosuppression’ at the heart of fatal COVID-19, in which, the observed NK cell exhaustion/depletion in severe COVID-19 could be a consequence of an overzealous IL15 storm, leading to their senescence and apoptosis.”
Thus, this study provides a valuable framework for understanding how the disease develops. The findings “not only pinpointed the precise nature of the cytokine storm, the culprit cell types and the organs, but also revealed disease pathophysiology, and helped formulate specific therapeutic goals for reducing disease severity.”
Further refining of the ViP signature through repeated filtering using newer and larger COVID-19 datasets may make it more precise and reliable. Even now, however, it can be used to inform therapeutic tools and to lay out a screening plan for potential therapeutic agents, not only in COVID-19 but also other viral pandemics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources