Researchers in the United States have shown that an engineered soluble decoy receptor tightly binds the spike protein of severe acute respiratory syndrome (SARS)-associated viruses. This could potentially provide protection against zoonotic betacoronaviruses such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent responsible for the current coronavirus disease 2019 (COVID-19) pandemic.
The engineered receptor, called sACE22.v2.4, served as an effective decoy that competed for the receptor-binding domain (RBD) on the spike protein. The spike protein is the surface structure these viruses use to bind wildtype angiotensin-converting enzyme 2 (ACE2), the human host cell receptor that enables viral entry.
Furthermore, the team from the University of Illinois at Urbana-Champaign and Orthogonal Biologics Inc. failed to find mutations within the spike RBD that could discriminate between an engineered decoy and wild type ACE2 receptors.
This suggests that resistance to engineered decoys will be rare and that decoys may be active against future outbreaks of SARS-associated betacoronaviruses, say the researchers.
A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review.
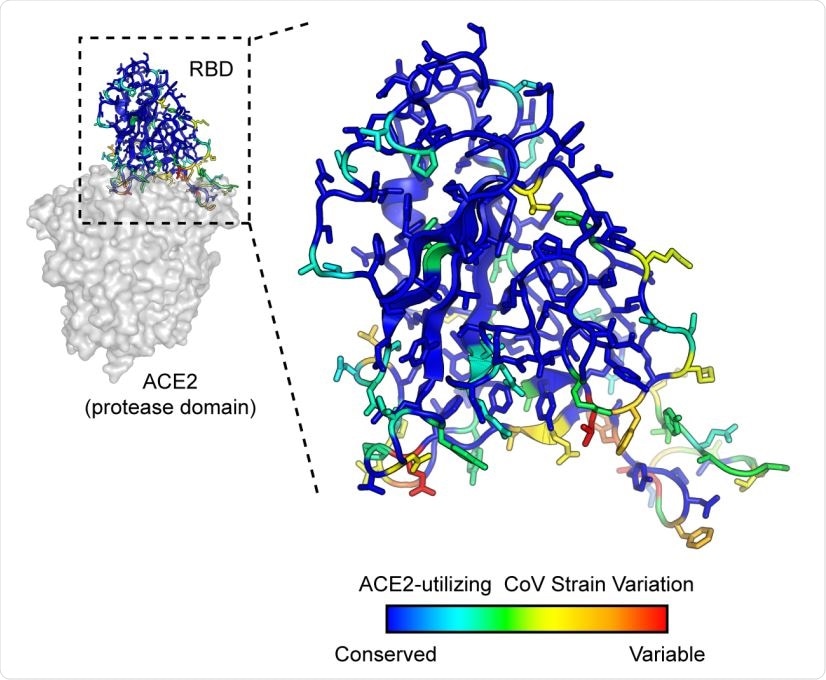
SARS-associated coronaviruses have high sequence diversity at the ACE2-binding site. The RBD of SARS-CoV-2 (PDB 6M17) is colored by diversity between 7 SARS-associated CoV strains (blue, conserved; red, variable).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Wild animals will almost certainly continue to be a source of future outbreaks
Zoonotic coronaviruses have spilled over from animal reservoirs several times over the last two decades, and it is almost inevitable that wild animals will continue to be a source of outbreaks in the future.
As SARS-CoV-2 continues to spread across the globe, infecting increasing numbers of people, the virus could potentially mutate and undergo genetic drift and recombination. Coronaviruses mutate at a moderate-to-high rate and recombination events have led to large changes in coronavirus genomes, especially among bats where levels of co-infection can be high.
This could have profound implications for the trajectory of the current COVID-19 pandemic, as well as the potential for future coronavirus pandemics and the prevalence of drug-resistant SARS-CoV-2.
Viral spike protein a popular target in drug development
The viral spike protein is a popular target for the development of neutralizing monoclonal antibodies, a number of which are currently in various stages of clinical trials.
However, in vitro studies have shown that the spike protein quickly develops escape mutations to all antibodies tested. Such mutations in spike retain high expression and affinity for ACE2 but are no longer recognized and bound by monoclonal antibodies.
"This has motivated the development of cocktails of non-competing monoclonals… to limit the possibilities for the virus to escape," said Erik Procko (University of Illinois) and colleagues. "However, even the use of monoclonal cocktails does not address future coronavirus spillovers from wild animals that may be antigenically distinct."
An alternative approach to monoclonal antibodies is to use engineered soluble ACE2 (sACE2) as a decoy to compete for the spike RBD.
"In principle, the virus has limited potential to escape sACE2-mediated neutralization without simultaneously decreasing affinity for the native ACE2 receptor, rendering the virus less virulent," says the team.
A combination of three mutations increased affinity for spike
Recently, the researchers identified a large number of mutations across the interface and distal sites of ACE2 that increase affinity for the viral spike protein. A combination of three of these mutations - called sACE22.v2.4 - increases the affinity for spike by 35 times and binds to the spike of SARS-CoV-2 with a comparable affinity to that of some of the most effective monoclonal antibodies.
However, the presence of mutations at or near to the interaction surface of such engineered decoy receptors provides an opportunity for spike variants to discriminate in favor of wild type ACE2, adds the team.
Testing this hypothesis
Now, Procko and colleagues have tested this hypothesis and shown that sACE22.v2.4 tightly binds to the spike RBDs of diverse SARS-associated betacoronaviruses from humans and bats, despite the ACE-2 binding site being a region with high sequence diversity.
In a comprehensive screen of all substitutions within the RBD, no mutations in the SARS-CoV-2 spike that confer high specificity for wild type ACE2 were found.
"The engineered decoy receptor is therefore broad against zoonotic ACE2-utilizing coronaviruses that may spill over from animal reservoirs in the future and against variants of SARS-CoV-2 that may arise as the current COVID-19 pandemic rages on," write the researchers. "We conclude that resistance to engineered decoys will be rare and that decoys may be active against future outbreaks of SARS-associated betacoronaviruses."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources