It is crucial to establish an effective and successful treatment strategy to mitigate this coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Favipiravir, a drug, has been used to treat influenza to date and investigated to treat life-threatening pathogens such as Ebola virus, Lassa virus, and now COVID-19.
Favipiravir is a pyrazinecarboxamide derivative that acts against RNA viruses. It targets the RNA-dependent RNA polymerase (RdRp) enzymes necessary for the transcription and replication of viral genomes.
In a recent bioRxiv* preprint publication, Christopher J. Russo et al. report the structure of favipiravir ribonucleoside triphosphate (favipiravir-RTP) in complex with the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) bound to a template: primer RNA duplex. They determine the structure by electron cryomicroscopy (cryoEM) to a resolution of 2.5 Å.
This is an important study for the design and development of novel antiviral therapeutics. The researchers observed that favipiravir-RTP is an inefficient substrate for viral RdRp. They propose that this is a consequence of a catalytically unproductive conformation adopted by the drug in the polymerase active site.
The SARS-CoV-2 viral RNA-dependent RNA polymerase is a promising drug target for the treatment of COVID-19. While nucleoside analogs, including remdesivir, sofosbuvir, and favipiravir, have shown a broad spectrum of activity against viral polymerases, current treatment regimens appear to have little or no effect on hospitalized COVID-19 patients (measured by overall mortality, initiation of ventilation, and duration of hospital stays). Insights into the mechanism at the molecular and structural levels become crucial.
The coronaviruses’ multi-subunit RNA synthesis machinery is a complex of non-structural proteins (nsp). This study shows the structure of the SARS-CoV-2 RdRp, comprising subunits nsp7, nsp8, and nsp12, in complex with template: primer double-stranded RNA and favipiravir ribonucleoside triphosphate (favipiravir-RTP).
The researchers first verified the assembled nsp7-nsp8-nsp12 SARS-CoV-2 RdRp complexes' activity, using primer extension activity assays. They then investigated the enzyme's ability to incorporate favipiravir-RTP: favipiravir-RTP appears to be a comparably poor substrate compared to the natural ribonucleotide triphosphates (rNTPs). Despite the low incorporation of favipiravir-RTP observed in primer extension assays, the researchers observe that it suppresses the completion of RNA replication at all concentrations tested, even in the presence of rNTPs. The inhibition was observed even when rNTPs are present at a considerable excess over the inhibitor.
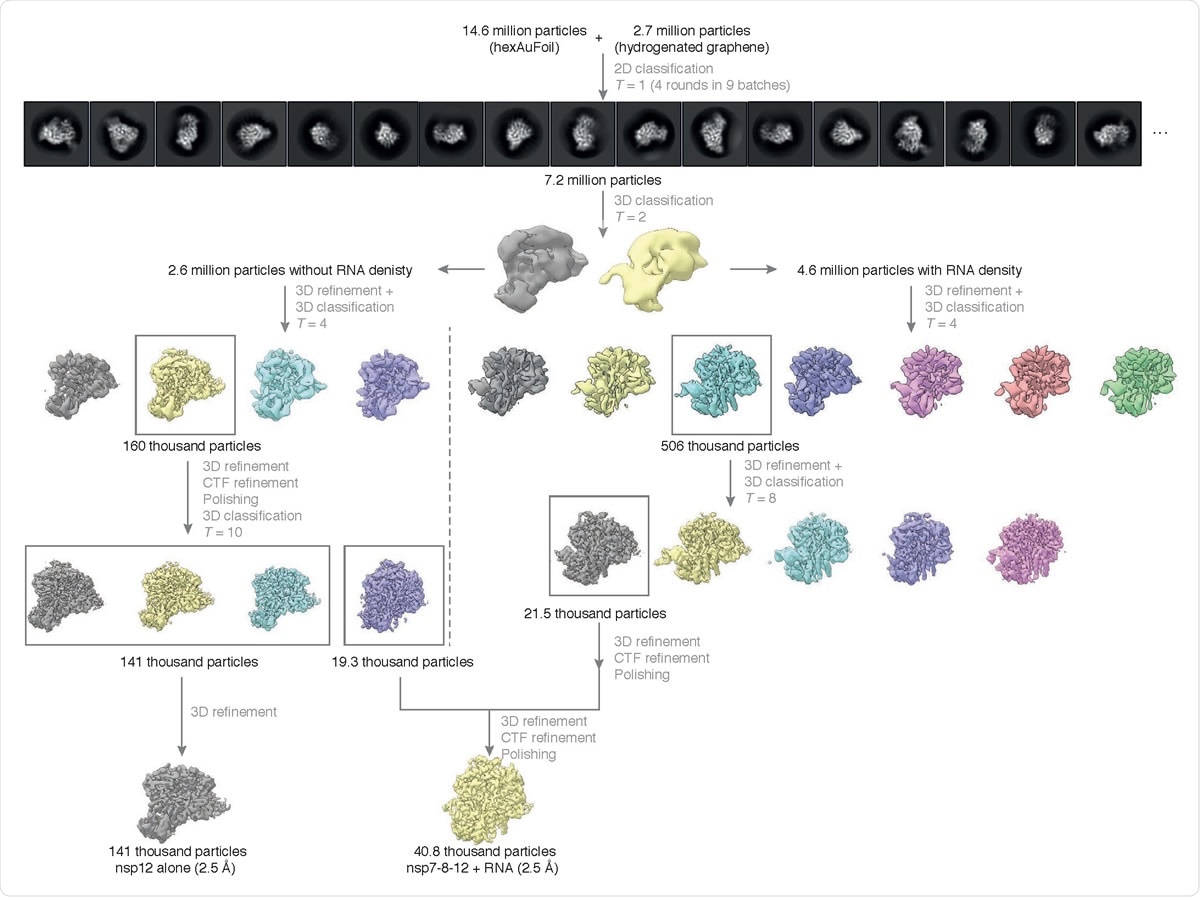
Cryo-EM data processing summary. The flowchart shows the main steps in the data processing, from particle picking, through classification, to final maps. A selected subset of the initial reference-free 2D class averages and all the intermediate 3D class averages computed during the processing of this dataset are shown. All 3D class averages, selected for subsequent rounds of processing, are boxed in gray, and the number of particles in each of these is shown. Further attempts to process the discarded classes are omitted from this chart for clarity, as these data did not contribute to the final particle set.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The researchers then proceeded to investigate the structural basis of this activity using cryoEM. This study elaborates on the methodologies, especially the challenges in cryoEM specimen preparation, including the further improvements the researchers made here.
The polymerase complex (comprising one nsp12, one nsp7, two nsp8 subunits, and a template: primer RNA) observed here is nearly identical to previously described reports. The inhibitor binding to the polymerase is non-covalent, with little to no covalent incorporation.
The researchers suggest that the observed non-productive configuration of favipiravir-RTP is promoted by an intrinsic feature of the favipiravir moiety. Possibly water-mediated hydrogen bonds between the drug and the triphosphate exist - although this is not unambiguously identified in this study. In support of this proposed mechanism, the researchers suggest that the T-1105-RTP (which lacks the fluorine and hence no possibility of hydrogen bond formation) is a more efficient substrate than favipiravir-RTP for SARS-CoV-2 RdRp
In conclusion, the researchers present the 2.5 Å resolution structure of the favipiravir-RTP bound to SARS-CoV-2 RdRp, using cryo-electron microscopy. This structure illuminates the coronavirus core RNA-synthesis machinery assembly, providing key insights at the molecular level.
This study throws light on the influence of this nucleotide analog inhibitor on RNA synthesis in vitro. Based on the structure reported in this study, the researchers discuss at least three possible directions for further efforts towards drug design against the SARS-CoV-2 RdRp.
The researchers admit that the cryoEM structure determination of this RdRp-RNA complex in combination with different inhibitors remains a challenge as a routine task. However, such studies will aid rational drug design and development for COVID-19 treatment.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Structural basis for the inhibition of the SARS-CoV-2 RNA-dependent RNA polymerase by favipiravir-RTP, Katerina Naydenova, Kyle W Muir, Long-Fei Wu, Ziguo Zhang, Francesca Coscia, Mathew J Peet, Pablo Castro-Hartmann, Pu Qian, Kasim Sader, Kyle Dent, Dari Kimanius, John D Sutherland, Jan Lowe, David Barford, Christopher J Russo, bioRxiv 2020.10.21.347690; doi: https://doi.org/10.1101/2020.10.21.347690, https://www.biorxiv.org/content/10.1101/2020.10.21.347690v1
- Peer reviewed and published scientific report.
Naydenova, Katerina, Kyle W. Muir, Long-Fei Wu, Ziguo Zhang, Francesca Coscia, Mathew J. Peet, Pablo Castro-Hartmann, et al. 2021. “Structure of the SARS-CoV-2 RNA-Dependent RNA Polymerase in the Presence of Favipiravir-RTP.” Proceedings of the National Academy of Sciences 118 (7): e2021946118. https://doi.org/10.1073/pnas.2021946118. https://www.pnas.org/doi/full/10.1073/pnas.2021946118.