The ongoing coronavirus disease (COVID-19) pandemic is caused by the SARS-CoV-2, which uses RBD of viral S glycoprotein (S protein) to bind to the angiotensin-converting enzyme 2 (ACE2) receptor with an affinity in the low nanomolar range. Accordingly, S protein is the prime target for antibody response against the virus.
Typical antibodies have antigen-binding co-determined by the variable domains of both its heavy chain (VH) and light chain (VL/VK); however, camelids produce unconventional heavy-chain-only antibodies that bind to antigens merely based on the variable domain of their heavy chain – the VHH domain (also known as nanobodies).
These VHHs are increasingly being used as functional antibody domains due to their small size and high stability, and VHH libraries have been screened for binders by phage and yeast display with significant success.
To leverage all the advantages of cell-free display, the research group led by Dr. Xun Chen (from the Broad Institute of MIT and Harvard) developed Cell-free VHH Identification using Clustering Analysis (or CeVICA), which is a novel, integrated platform for in vitro VHH domain antibody engineering.
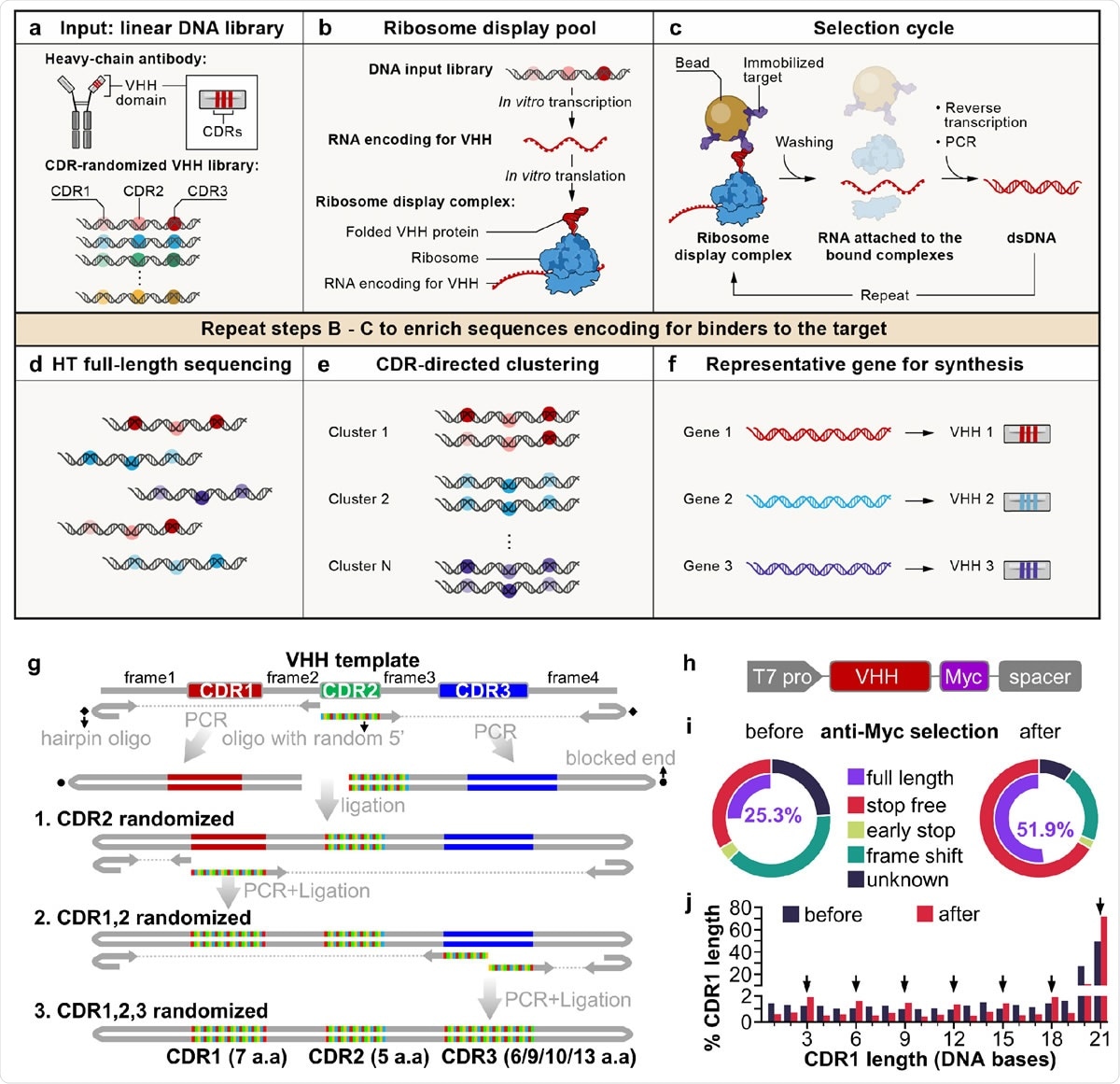
A cell-free antibody engineering platform for rapid isolation of antibodies from large synthetic libraries. (a) The workflow takes linear DNA library as input. (b) Ribosome display links genotype (RNAs transcribed from DNA input library that are stop codon free, and stall ribosome at the end of the transcript) and phenotype (folded VHH protein tethered to ribosomes due to the lack of stop codon in the RNA). (c) Selection cycle enriches DNA encoding for VHHs that binds immobilized targets. (d) High throughput sequencing of full-length VHHs. (e) Sequences are grouped into clusters based on the similarity of their CDRs, each cluster is distinct and represents a unique binding family. (f) The system outputs one representative sequence from each cluster to be synthesized and characterized for specific downstream applications. (g) Workflow for generating VHH library. VHH CDR randomization was introduced by PCR using a hairpin oligo (blocks DNA end from ligation) and an oligo with a random 5’ sequence, followed by orientation-controlled ligation. Three successive PCR plus ligation cycles randomizes all three CDRs. (h) The final DNA library sequence structure. (i) One round of ribosome display and anti-Myc selection was performed after randomization of CDR1 and CDR2. The pie chart shows percentage of indicated sequence categories before and after anti-Myc selection. (j) Length distribution of DNA region encoding CDR1 of the VHH library before and after anti-Myc selection. Arrows indicate all correct-frame lengths showing increased percentage after anti-Myc selection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Optimized design
In a nutshell, this platform combines a completely new design and generation method for randomized VHH libraries, optimized ribosome display, and selection cycle, as well as a computational approach to carry out global binder prediction from post-selection libraries.
To adequately perform affinity maturation, which represents a critical stage in antibody development in animals, the researchers have designed and performed an affinity maturation strategy based on CeVICA to expand the affinity of RBD-binding VHHs.
Subsequently, they have calculated their amino acid profiles in regards with the position, and determined the change in each amino acid proportion at each position for each VHH – generating, in turn, a percent point change table.
The researchers have also defined putative beneficial mutations and assembled a list of them for each VHH, and incorporated their various combinations into each VHH parental sequence in order to generate manifold mutated variants for the final assessment.
Finally, they have examined the potential impact that these VHH sequences may have on immunogenicity in humans since a big concern linked to the therapeutic use of VHH antibodies is the possibility that – as camelid proteins – they would prompt an immune response.
Efficient and meticulous binder prediction
Among 14 experimentally-tested binders, six of them showed inhibition of the infection with SARS-CoV-2 S pseudotyped lentivirus. Antibody affinity maturation also increased binding affinity, as well as the potency of inhibition.
Moreover, a distinctive capability of CeVICA for efficient and exhaustive binder prediction enabled retrospective validation of the fitness of the synthetic VHH library design shown in this study as well as revealing direction for future refinement.
The obtained data also imply that at least some of the VHH hallmark residues may be converted to human residues without losing binding fitness. These conversions may then serve as frame features of future VHH library designs, and subsequently enhance tolerance in vitro engineered VHHs by humans
A viable strategy for refining antibodies
Taking everything into account, the extension of CeVICA for affinity maturation provides a viable strategy for refining antibody function, while supplementary iterations of the affinity maturation process may yield additional enhancement of antibody properties.
"Using CeVICA, we generated a large collection of antibodies that can bind the RBD domain of the SARS-CoV-2 spike protein and can neutralize pseudotyped virus infection, thus providing an important resource", study authors accentuate in this bioRxiv paper.
In any case, such a harmoniously integrated procedure opens the door for automation and could provide an important tool for rapid, scalable and reliable antibody generation. A CeVICA technology framework may overcome limitations of in vivo fitness of laboratory-generated antibodies, but also overall efficiency, which needs to be substantiated by further studies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources