Coronavirus disease 2019 (COVID-19) is known for the diversity of its clinical features, ranging from asymptomatic infection to lethal multi-system breakdown. The risk factors for severe disease include advanced age as well as chronic underlying diseases.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Angiotensin-converting enzyme 2 and inflammation
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen that causes COVID-19, gains entry to host cells by binding to the angiotensin-converting enzyme 2 (ACE2), a receptor expressed on a variety of cells. This molecule is associated with many inflammatory and metabolic events and pathways, including cardiovascular disease (CVD) and the renin-angiotensin system (RAS). It also plays a part in regulating the balance between various amino acids.
Inflammation hypothesis
The metabolic and inflammatory disease conditions that enhance the risk of severe COVID-19 all have a common inflammatory basis, which is traceable to a dysregulated metabolic process.
NMR spectroscopy
The study was aimed at eventually improving the current understanding of risk factors for severe COVID-19, the systemic effects of the infection and residual cardiac and metabolic disturbances.
The study, which was conducted by a group of scientists based in Belgium, used blood samples taken from three groups of patients, two cross-sectional and the third using repeated sampling over time. There were 250 metabolites, including ratios, measured by nuclear magnetic resonance spectroscopy. This technology was used because it allows many metrics to be evaluated at the same time, while providing the required accuracy of resolution to understand variations in the lipid and lipoprotein profile.
The biomarkers included inflammatory markers such as the acetylation of glycoproteins, amino acid and lipoprotein particle concentrations, and lipid levels. These were then evaluated with respect to their link to the severity of the disease.
GlycA: marker of inflammation
Glycoprotein acetylation (GlycA) is a new systemic inflammatory marker, found at higher levels in CVD, type II diabetes and an elevated body mass index. This composite marker comprises the signal from the N-acetyl group on a pool of circulating glycoproteins.
The dynamic GlycA signal is more accurate in reflecting inflammation than the more commonly used CRP, because it is more durable over time and reflects multiple sites and inflammation levels. It can indicate inflammation even when the CRP is negative. The GlycA actually condenses the baseline chronic inflammatory burden. This is especially valuable since severe COVID-19 is associated with low-grade inflammation, and with an elevated risk of CVD during the episode and after recovery.
Metabolic biomarkers for severe COVID-19
Raised GlycA, albumin and the GlycA/albumin ratio, and high creatinine, were strongly linked with severe COVID-19. Of course, these measures are core components of the ‘Infectious Disease Score’ (IDS), developed by Julkunen et al., a reliable composite measure of disease severity. This biomarker score is linked to the odds of developing severe pneumonia within the next eight years.
The study showed that a high IDS is independently associated with a fourfold rise in the risk of severe COVID-19. The amino acids phenylalanine and leucine are also linked to severe disease. Valine and branched-chain amino acid residues were associated with severity in only one of the two cohorts. Similarly, glutamine (Gln), tyrosine (Tyr) and Histidine (His) residues also showed association in one cohort only.
COVID-19 severity was inversely linked to higher lipoprotein (LP) levels. Higher cholesterol content and absolute phospholipid content within particles were markers for less severe disease in most LP classes, except for VLDL and large HDL. On the other hand, higher triglyceride content in IDL and LDL was a marker for greater severity, as was the ratio of phospholipids to total lipids within HDL, IDL and LDL. The ratios of other lipoprotein components to total lipids was otherwise proportional to the disease severity, as was their absolute levels.
Increased polyunsaturated fatty acid (PUFA) and monounsaturated fatty acid (MUFA) content were linked to lesser and greater severity, respectively. The strongest associations for severe disease were with linoleic acid and total omega-6 FA. In contrast, higher ratios of total omega-3 and docosahexaenoic acid (DHA) to total FA levels were associated with a 20% to 30% reduction in severity.
In the second part of the study, the association between 72 biomarkers and severe to critical COVID-19 was validated. These include serum albumin, GlycA/albumin and IDS, phenylalanine (alone among the amino acids), and the omega-6 and PU FAs, with the PUFA/MUFA ratio. Low admission HDL cholesterol in small HDL particles was characteristic of more severe cases.
Longitudinal trends
Samples taken during follow-up showed that some of these biomarkers exhibited worsening at day seven post-admission but returned to admission levels by discharge.
At 30 days from discharge, they were at baseline levels. However, choline, phosphatidylcholine and phosphoglyceride remained unchanged at day seven and at discharge but increased thereafter.
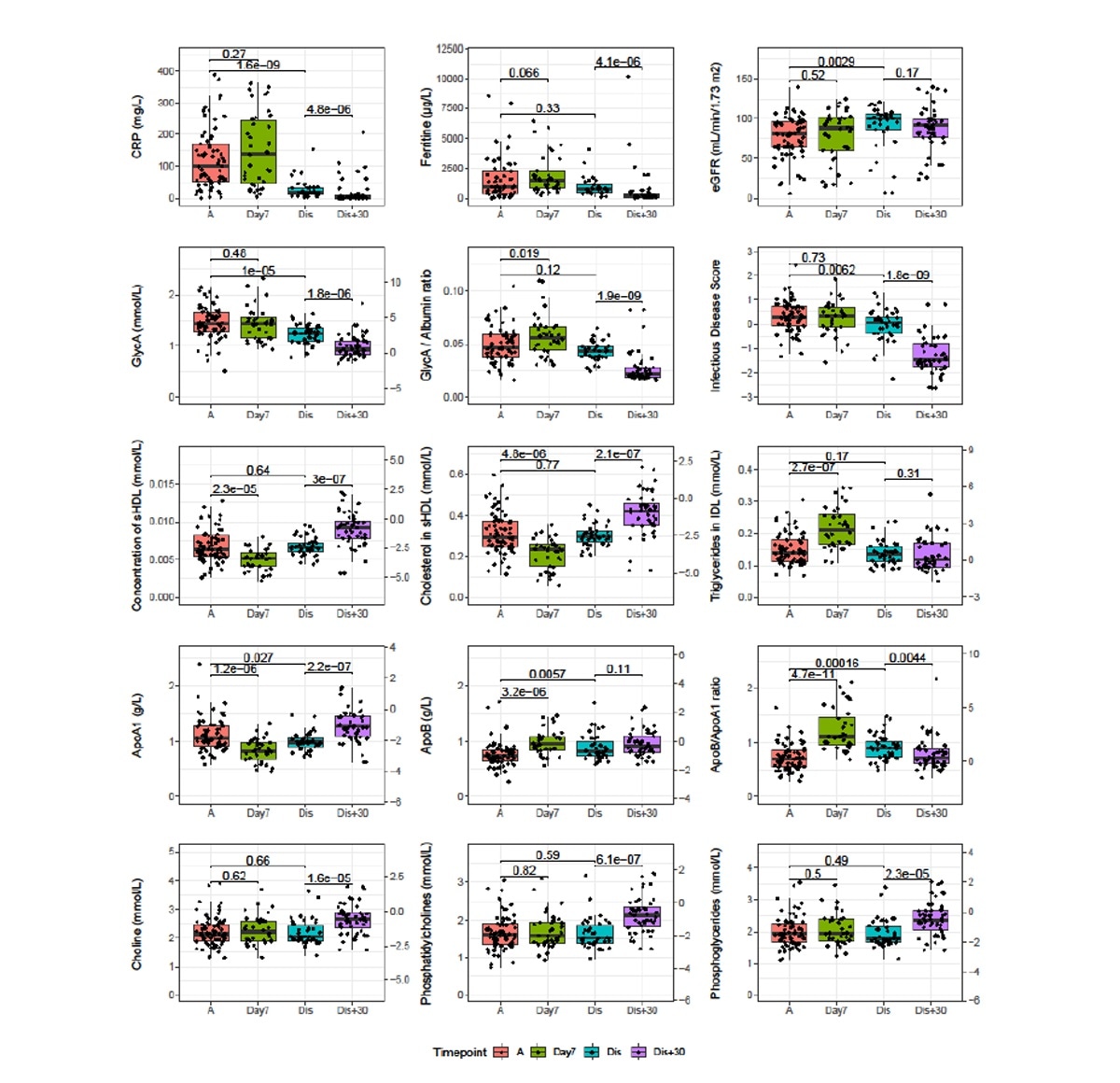
Longitudinal analysis of severe and critical COVID patients of the CONTAGIOUS cohort. Patients of the CONTAGIOUS cohort were sampled at COVID admission (A, red), 7 days post admission (Day7, green), time of hospital discharge (Dis, blue) and 30 days post hospital discharge It is made available under a CC-BY-NC-ND 4.0 International license . preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. medRxiv preprint doi: https://doi.org/10.1101/2020.11.09.20228221; this version posted November 12, 2020. The copyright holder for this (Dis+30, purple). Indicated are p-values of Mann-Whitney U tests comparing the indicated timepoints. Where depicted, the y-axis on the right side of the graph normalizes the data using the mean and standard deviations observed in Finnish demographic cohorts42. Full comparison results for all variables associated to severity are available in supplementary table S4. ApoA1: Apolipoprotein A1, ApoB: Apolipoprotein B, CRP: c-reactive protein, GlycA: glycoprotein acetylation.
Implications and future directions
In the study’s findings, the associations between these markers and disease severity corresponded at least as well as more traditional acute inflammatory markers. This metabolic profile indicates four important areas:
- The link between COVID-19 and inflammation
- The changes in energy requirements of the hyperinflammatory immune cells due to lower oxidative phosphorylation in the mitochondria. This may indicate an increased breakdown of glutamine and of branched-chain amino acids in mild cases, but not in severe cases where aerobic glycolysis takes over.
- The low incidence of severe COVID-19 in patients with phenylketonuria could be due to low phenylalanine levels
- The link between cholesterol within lipoprotein particles, the LP profile, as well as COVID-19 severity and the importance of serum FA levels in severe COVID-19, especially linoleic acid. This could be not just due to the effect of inflammation on the metabolic process, but an indication of a “genetic predisposition to severe infection,” as with HDL cholesterol. In fact, serum apoB, serum cholesterol, cholesterol/phospholipid content of LDL, IDL and VLDL are genetically associated with cytokine levels, and lower levels are linked to more severe disease. Again, linoleic acid binds to the viral spike protein, and higher levels could thus mediate a reduction in the severity of the disease.
The study concludes, “Our results point to systemic metabolic biomarkers for COVID-19 severity that make strong targets for further fundamental research into its pathology.”
The researchers point out that more studies on NMR markers in other infectious conditions like severe pneumonia are necessary to disentangle the causes and effects of observed biomarker changes in COVID-19 from others.
However, this study shows, “the metabolic fingerprint of COVID-19 severity consists of a pervasive inflammatory signature.” The nonspecific range of biomarker levels reflects the highly variable spectrum of clinical features in this illness, making further research essential to elucidate their role in the disease.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.