The current coronavirus disease 2019 (COVID-19) pandemic, caused by the SARS-CoV-2 agent, is widely believed to be of zoonotic origin (meaning it had originated in animals and has since jumped to human hosts). Much scientific research has subsequently been directed toward understanding the genetic diversity of RNA viruses, like SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
An animal model is useful for studying viral genetic variation in the host and between different hosts. SARS-CoV-2 animal models have included Syrian hamsters, ferrets, cats, and dogs, among which natural transmission has been seen only in mink, cats, and ferrets. Only in cats, however, has SARS-CoV-2 been detected in the lower respiratory tract; infection in this region for humans has been linked to severe cases of COVID-19.
Transmission bottleneck
The term transmission bottleneck refers to a significant reduction in virus population during transmission. Observing the development of transmission bottlenecks is important for understanding the evolution of respiratory viruses. Narrow transmission bottlenecks, where there is weak natural selection, could reduce adaptations in the seasonal flu virus and could also play a role in SARS-CoV-2 transmission. Accurate knowledge of the size of the SARS-CoV-2 transmission bottleneck could thus help forecast its evolution.
Since previous studies in humans on the transmission bottleneck have reported conflicting results, a new study has utilized a cat model. The study's findings are available on bioRxiv*, the preprint server
SARS-CoV-2 genetic diversity in cats
The researchers, from the University of Wisconsin-Madison and Emory University in the USA, used genome sequencing of the virus in index cats, cats first infected with the virus and in other cats that the index cats transmitted the virus to. They then studied changes in these sequences over time to determine genetic variation.
They inoculated three domestic cats with a SARS-CoV-2 human isolate from Tokyo. Each inoculated cat was then kept with a virus-free cat, starting a day after the inoculation. They then collected nasal swabs daily for up to 10 days.
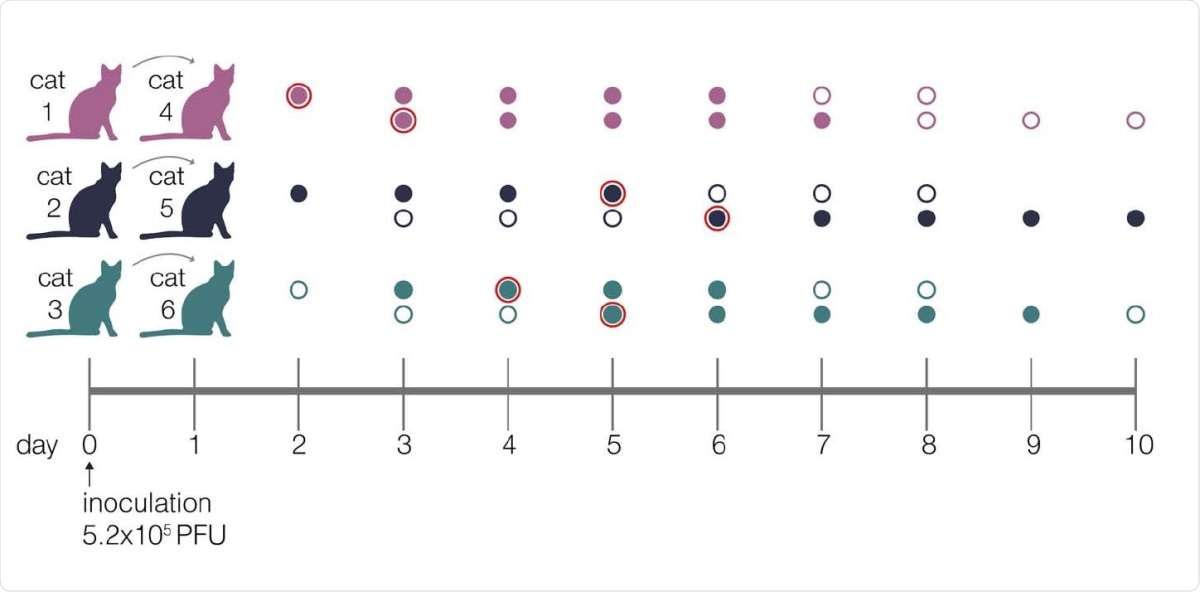
Experimental timeline. Schematic representing the sampling timeline for the three transmission pairs. Index cats were inoculated on day 0 with 5.2e5 PFU of a human isolate (hCoV-19/Japan/UT-NCGM02/2020) and were co-housed with a naive cat starting on day 1. Within each transmission pair, the top row of circles represent the index cat and the bottom row represents the contact cat. Open circles represent days on which there was no detectable infectious virus as indicated by plaque assay, and closed circles highlight days when live virus was recovered. Circles with a red outline indicate timepoints which were used in the betabinomal estimate to calculate transmission bottleneck sizes.
They found 86% of the variants were found in a single cat, 8% in 2 to 5 cats, and the remaining variants were found in all six cats. Around 43% of the mutations occurred at a frequency of 3 to 10%, and about 8% occurred at a frequency of 40-50%, with the frequency of variations remaining about the same over time in the index cats.
Purifying selection is when harmful mutations are removed and result in more low-frequency variants. Positive selection results in more high-frequency variations and is when useful variants are retained in the population. Since about half of the variants occur at low frequency, purifying selection is predominant in cats. By comparing nonsynonymous and synonymous nucleotide diversity, the authors found that genetic variations in the index cats were driven by genetic drift, or mutations that occurred by random chance.
To determine the transmission bottleneck size in cats, the authors studied how much genetic diversity was lost after transmission. They found a lower number of variants in the contact cats compared to its index cat. Furthermore, they found the effective bottleneck size to be five (after two and three days, index and contact cat, respectively); three (after five and six days); and two (after four and five days). This is similar to other studies on flu virus transmission in humans.
SARS-CoV-2 mutation rate in cats is slow
Although genetic variation within hosts was low, there were two notable mutations. One, the S H655Y, variant has been seen before and is related to the spike protein glycoprotein fusion. The other variant, E S67S, has not been seen before. Both variants were transmitted in two of the three cats.
The results suggest that genetic selection within hosts is weak and the transmission bottleneck is narrow, even during close contact, indicating that SARS-CoV-2 is perhaps already well adapted to mammalian hosts.
"The strong role of genetic drift may combine with the relatively slow mutation rate and narrow transmission bottlenecks to slow the overall pace of viral evolution," write the authors.
The narrow transmission bottleneck finding conflicts with some human studies. However, the authors suggest that the current study's experimental design avoids challenges faced in previous human studies; namely, the potential for other sources of exposure to infection, and not only intra-household transmission.
There have been reports of natural transmission from humans to cats, so cats could be a feasible model for studying virus genetic diversity. The narrow transmission bottleneck suggests that SARS-CoV-2 variations that could affect innate immunity or antivirals could be slow.
However, variants do arise early and are capable of being transmitted in cats, a potential reservoir species, and host-specific adaptations are also inevitable in SARS-CoV-2. This may also affect humans if exposed to species-specific adaptations, requiring continued studies on sequencing and understanding SARS-CoV-2 genetic diversity in different hosts.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Braun, K. M. et al. (2020) Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. bioRxiv. https://doi.org/10.1101/2020.11.16.384917, https://www.biorxiv.org/content/10.1101/2020.11.16.384917v1
- Peer reviewed and published scientific report.
Braun, Katarina M., Gage K. Moreno, Peter J. Halfmann, Emma B. Hodcroft, David A. Baker, Emma C. Boehm, Andrea M. Weiler, et al. 2021. “Transmission of SARS-CoV-2 in Domestic Cats Imposes a Narrow Bottleneck.” Edited by Andrew Pekosz. PLOS Pathogens 17 (2): e1009373. https://doi.org/10.1371/journal.ppat.1009373. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009373.