The coronavirus disease 2019 (COVID-19) pandemic emerged in late-2019, with the finding of several cases of a novel pneumonic virus in China's Wuhan City. This was initially called the novel coronavirus, but its final designation became severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It is thought by many researchers to be phylogenetically very close to the coronaviruses infecting Chinese horseshoe bats and perhaps the Malayan pangolin, but the exact chain of animal-to-human transmission remains elusive.
About miRNAs
MicroRNAs, or miRNAs, are molecules that are important in deciding what happens after a gene is copied from the DNA to RNA – should it proceed to the production of the protein encoded, and if so, when and how? These questions are answered by post-transcriptional gene regulation, and miRNAs help answer them. Thus, these tiny molecules affect multiple biological processes across a variety of cell types.
Several miRNAs are linked to host-pathogen interactions, and may be linked to cross-acting host-viral interplays following transcription that drives the clinical course of the illness. It is important to understand viral miRNAs and their targets in the human genome to identify therapeutic strategies.
The current study, published in the journal Bioinformatics in November 2020, describes the efforts made to identify precursor miRNA-like (pre-miRNA) molecules in the SARS-CoV-2 genome. Using infected epithelial cells in culture, they found six pre-miRNAs, which might be the precursors of 8 mature miRNAs built up in the cell during viral infection.
The researchers used sequences from the NCBI, both the viral genome and its expression in Calu-3 cell lines infected with the virus. They then used extracted structural features for their deep learning analysis by three independent methods; namely, a deep convolutional neural network (CNN), the deeSOM model and a one-class support vector machine (OC-SVM).
These helped to predict pre-miRNAs, and those with the top scores were identified. This was followed by looking for genes that were expressed at a significantly higher or lower level in infected cells compared to healthy cells – differentially expressed genes (DEGs) – and for cellular metabolic pathways running at higher levels in infected cells.
The researchers found six reliable pre-miRNA precursors predicted by the algorithms to be encoded in the viral RNA. These would give rise to eight mature viral miRNAs that could potentially regulate host genes. Each miRNA was found to arise at a different time point following viral infection of the cell. These viral miRNAs were distinct from human miRNAs or those from any other organism except the fruit fly.
Their potential human targets were then predicted using Diana (MR-microT software) and miRDB software set for custom prediction. When only those targets with a score over 70 were considered, they found 725 potential targets, with a mean of around 100 targets for each mature miRNA.
Among these, about 100 potential human genomic targets were predicted to be affected by the six predicted potential viral pre-miRNAs. Of these, 28 were downregulated, which agrees with the hypothesis that predicted mature miRNAs silence many host genes.
When the scientists explored the functional pathways expressed at higher levels, they found that the development of neurons and dendrite formation was especially targeted. This could perhaps explain why COVID-19 often produces neurological symptoms.
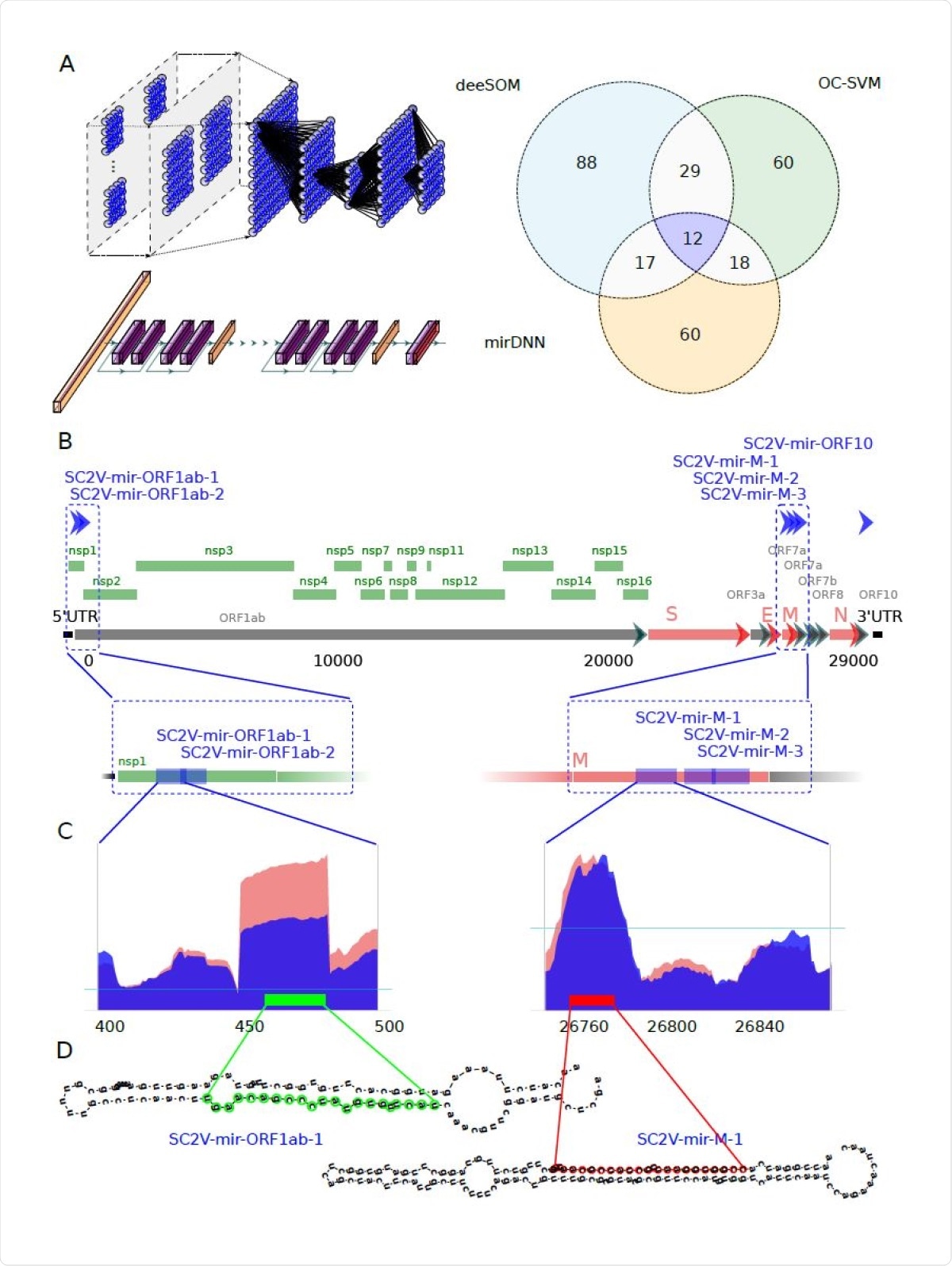
Identification of potential miRNA from six precursors hidden in the protein-coding genome of SARS-CoV-2. A) Venn diagram showing the number of candidate sequences to pre-miRNAs in the SARSCoV- 2 virus found by three machine learning methods: deeSOM (blue), mirDNN (orange) and OC-SVM (green). Deep models are schematically represented. On top is the deeSOM, several layers with ensembles of self-organizing maps, with elastic map size and automatic depth. On bottom is the mirDNN, a novel convolutional neural network with several layers and identity blocks. B) Genome location of the six predicted pre-miRNAs in the SARS-CoV-2 virus. C) Two examples of the read profiles in small RNA-seq samples of Calu-3 cell cultures at 24 hours upon infection. The horizontal blue line indicates the average reads counts for the whole genome. D) Predicted hairpin structures of the two SARS-CoV-2 pre-miRNAs shown in C).
Mutations in miRNA and the species barrier
The comparison of the miRNAs with those in other phylogenetically related coronaviruses showed that some recent mutations had taken place to give rise to the predicted novel mature miRNAs, which could have promoted zoonotic transfer.
A very interesting finding was that all the six viral pre-miRNAs overlapped with protein-encoding genes on the open reading frames (ORFs). These were found to be conserved in the bat viral genome but not the pangolin genome. The mutations between the bat genome and SARS-CoV-2 spared the protein-encoding genes, though point mutations were found in the seed sequences of three of the predicted mature miRNAs. This could influence the way these miRNAs recognize the host mRNA targets.
Seven of the eight mature miRNAs also showed changes between the pangolin and SARS-CoV-2, but only one altering the protein-coding sequence. It is possible that the point mutations in the spike RBD from pangolin and bat coronaviruses could have enhanced SARS-CoV-2 binding to human angiotensin-converting enzyme 2 (ACE2) and facilitated rapid viral transmission between humans.
These were not found to alter the protein encoded, but did affect the potential effects of these newly derived miRNAs on the type and number of genes transcribed in the infected cell. Overall, the findings led the researchers to suppose that these novel miRNAs that potentially target human genes may be a genomic feature that facilitated the species-crossing leap from the intermediate animal host into humans.
It is still unknown how cytoplasmic RNA viruses can generate miRNAs, since their production is classically considered to be initiated in the nucleus, and is dependent on nuclear enzymes. In the case of SARS-CoV-2, some viral genomic proteins could play a part in cytoplasmic miRNA production within the host cell. These include nsp15, an endoribonuclease, and nsp1, a cytoplasmic endonuclease that can disrupt the nuclear pore, allowing nuclear contents to enter the cytosol.
Most human miRNA genes are in non-coding regions, but some are in the exons or intron-exon junctions. The exonic miRNAs may be processed and may potentially destabilize the parent gene transcript. The overlap between the eight potentially active miRNAs discovered in the current study with protein-coding genes is intriguing in this context.
Some have postulated that the processing of miRNAs encoded in an RNA virus within a human cell wipes out the viral genome itself and thus prevents viral replication. Conversely, others estimate that enough miRNAs are produced once less than 1% of viral replication occurs in the infected cell, allowing viral replication to proceed unhindered.
What are the implications?
The role of miRNAs has been a matter of speculation, but many think that they modulate viral replication because of their potential effects on host responses and viral transcription. In fact, many of the silenced target genes are found to be involved in respiratory diseases and antiviral responses.
RNA-sequencing methods could miss the effects of many viral miRNAs on the host targets because up to 40% of human genes targeted by miRNAs can be silenced, stopping the production of their encoded proteins without any observable shift in RNA levels.
The current study suggests that proteins may either exert negative feedback on their encoding genes via regulatory loops, or that viral miRNAs may be produced at different points during viral replication or in a viral charge-dependent manner. This could induce gene silencing in certain situations. If so, circulating viral miRNAs could serve as biomarkers of COVID-19 progression.
The researchers comment, "Collectively, our observations suggest that SARS-CoV-2-derived miRNA-like sequences may modulate the host transcriptome promoting the infection progression." If so, understanding more about how these miRNAs act, how they promoted the emergence of the novel coronavirus, and how they affect the human host cell, will help to discover effective new therapeutic strategies.