The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has profoundly affected public health globally, leading to an unprecedented socio-economic crisis. Coronavirus disease 2019 (COVID-19) manifests as a respiratory illness, which disproportionately affects the elderly population and people with comorbidities. Patients with severe COVID-19 developed respiratory failure caused by poor gas exchange and massive lung inflammation.
This novel virus is genetically 79.5% similar to SARS-CoV-1 and 50% similar to the Middle East respiratory syndrome (MERS). The SARS-CoV-2 virus also used the same host receptor - angiotensin-converting enzyme 2 (ACE2) - as SARS-CoV-1 for entry into the host cell by binding its S protein to ACE2. The S protein binds ACE2 more strongly than SARS-CoV, though these 2 S proteins have similar tertiary structures.
SARS-CoV-2 activates the immune system of the host and causes widespread inflammation in patients with severe COVID-19. Knowing how the virus engages with the host immune system will enable the development of much needed therapeutic strategies to fight COVID-19.
Assessing the role of SARS-CoV-2 surface proteins in host immune response to the virus
Researchers from the Washington University School of Medicine, United States of America, recently demonstrated that the SARS-CoV-2 surface proteins alone triggered innate cell activity and the interferon signaling pathway. They showed that surface proteins spike (S) and envelope (E) of SARS-CoV-2 activate the key immune signaling interferon (IFN) pathway in both immune cells and epithelial cells independent of viral infection and replication both in vitro and in vivo. Their study has been published on the pre-print server bioRxiv*.
These surface proteins trigger the generation of reactive oxidative species and elevation in human and murine specific IFN-responsive cytokines and chemokines, similar to what happens in severe COVID-19 patients. Induction of IFN signaling relies on established but discrepant inflammatory signaling mediators as the activation induced by the surface protein S is dependent on IRF3, TBK1, and MYD88, while that of the envelope protein E is largely MYD88 independent.
We show that these viral antigens individually alter the expression of key chemokines and cytokines, including many regulated by IFN, in both human and murine cell lines.”
Moreover, these viral proteins, specifically the E protein, triggered peribronchial inflammation and pulmonary vasculitis in mouse models. Finally, the researchers showed that the organized inflammatory infiltrates rely on type I IFN signaling, specifically in the case of lung epithelial cells. These findings highlight the crucial role of SARS-CoV-2 surface proteins, especially the less known and less studied E protein, in triggering cell-specific inflammation and their potential for therapeutic intervention.
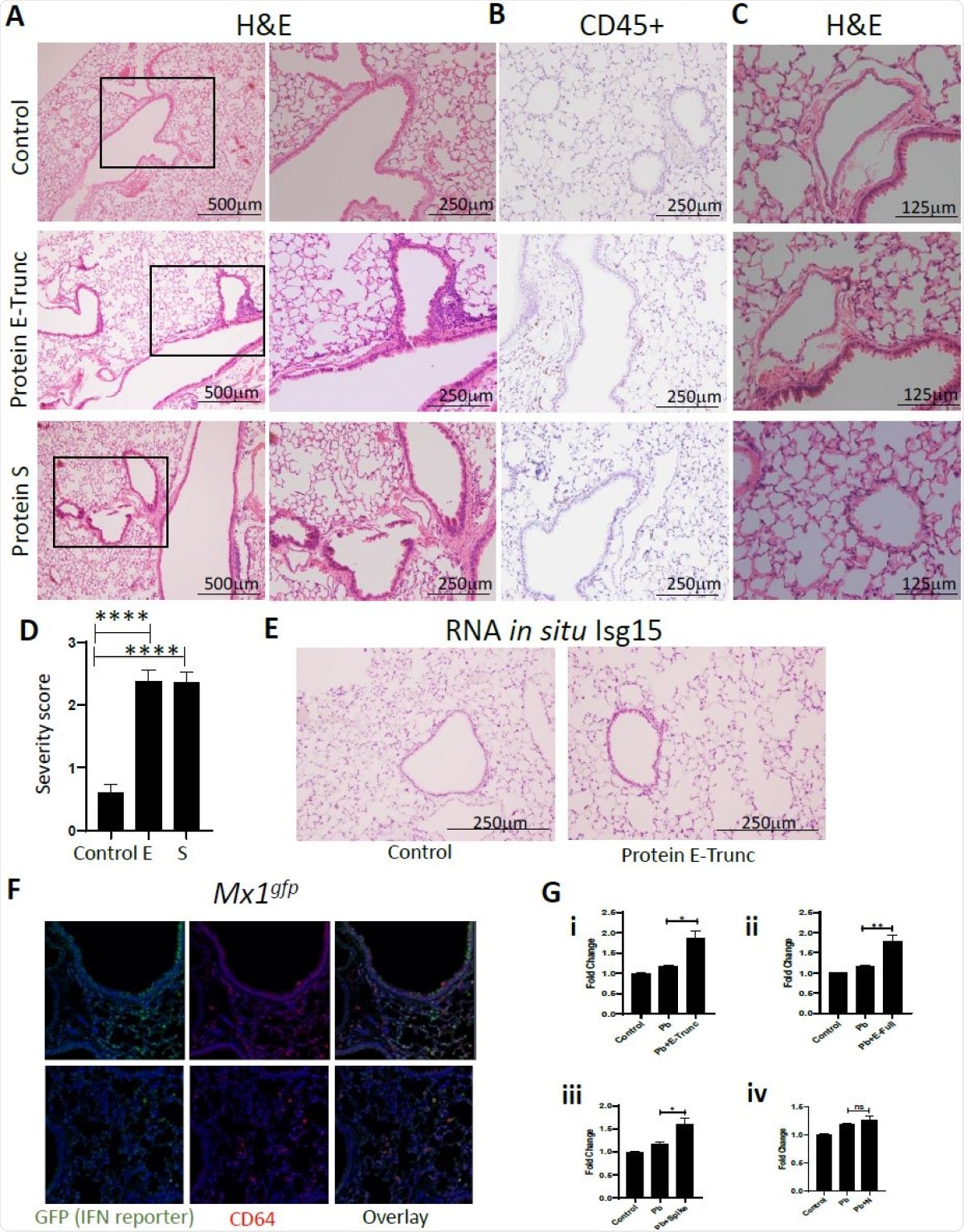
SARS-CoV-2 Protein E Induces Lung Inflammation and Vasculitis in Mice. (A) Representative images of lung cross-sections from mice after intranasal delivery of control or 10µg of E-Trunc and S. H+E stained sections are shown. Boxed areas on the left are magnified to the right. (B) Representative images of the lung cross-sections immunostained for CD45 expression. (C) Representative images of lung cross-sections focused on blood vessels in the above conditions. Scale bars depicted in each picture. (D) Quantification of percent of lobes with inflammatory infiltrates in lungs harvested in the above conditions. (n=3 mice per condition). (E) Representative images of the lung cross-sections stained for Isg15 by RNA in situ (n=3 mice per condition). (F) Representative immunofluorescent images of the lung cross-sections immunostained for GFP and CD64 expression 3 days after intranasal delivery of 10ug of E or control (n=2 mice). (G) Fold change in IFN reporter activities in A549 cells with treated with control, Pb at 10µg/ml and SARS-CoV-2 proteins (E-Trunc (i), E-Full length (ii), S (iii), N (iv)) individually at 2µg/ml with Pb at 10µg/ml for 24 hours. (n=2 experiments; 6 biological and 9-21 technical replicates with all proteins). Graphs depict average with SEM. *p<0.05 ,**p<0.01, ****p<0.0001 and ns denotes not statistically significant. Mann-Whitney used for statistical analysis.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Findings highlight the importance of the role of coronavirus surface proteins in severe COVID-19
The innate immune system of our body is our first line of defense critical for viral clearance while also playing a key role in the pathogenesis of many viral pathogens. Many studies have shown that COVID-19 pathogenesis is characterized by a hyperinflammatory state and an uncontrolled immune response. Patients with severe COVID-19 experience a “cytokine storm” due to the release of high levels of proinflammatory mediators, and this strength of this storm is directly proportional to the viral load and severity of disease.
Our observations also highlight the importance of the direct effect of coronavirus surface proteins and will usher investigation of other viral surface proteins as determinants of the host-pathogen interaction.”
This study showed that the viral antigens alone altered the expression of proinflammatory chemokines and cytokines, including the ones regulated by IFN, in both human and murine cell lines. The authors believe that their study has significant implications for the virus-host immune response as they show that the activation of innate immune signaling pathways happen independently of viral nucleic acid detection. A better understanding of the immune response to viral surface proteins is important for decoding how this novel virus engages with the immune system of the host.
Finally, this work has broad implications for the pathogen-host immune response; we show that activation of innate immune signaling pathways independent of viral nucleic acid detection by pathogen recognition receptors engage host immunity similarly to complete infectious virus.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Gautam Anand, Alexandra M. Perry, Celeste L. Cummings, Emma St. Raymond, Regina A. Clemens, Ashley L. Steed (2020) Surface proteins of SARS-CoV-2 drive airway epithelial cells to induce interferon-dependent inflammation. bioRxiv 2020.12.14.422710; doi: https://doi.org/10.1101/2020.12.14.422710, https://www.biorxiv.org/content/10.1101/2020.12.14.422710v1
- Peer reviewed and published scientific report.
Anand, Gautam, Alexandra M. Perry, Celeste L. Cummings, Emma St. Raymond, Regina A. Clemens, and Ashley L. Steed. 2021. “Surface Proteins of SARS-CoV-2 Drive Airway Epithelial Cells to Induce IFN-Dependent Inflammation.” The Journal of Immunology 206 (12): 3000–3009. https://doi.org/10.4049/jimmunol.2001407. https://journals.aai.org/jimmunol/article/206/12/3000/234392/Surface-Proteins-of-SARS-CoV-2-Drive-Airway.