As the coronavirus disease 2019 (COVID-19) pandemic continues to intensify in many parts of the world, vaccine development is proceeding apace. Indeed, the earliest approved vaccines are now being administered to hundreds of thousands of people the world over.
However, the occurrence of mutations leading to the emergence of multiple variants has led to uncertainty about the possibility of escape mutations, which would render these vaccines ineffective.
A new preprint on the bioRxiv* server, appearing in December 2020, indicates that these fears may be unnecessary, given the current state of knowledge about the virus.
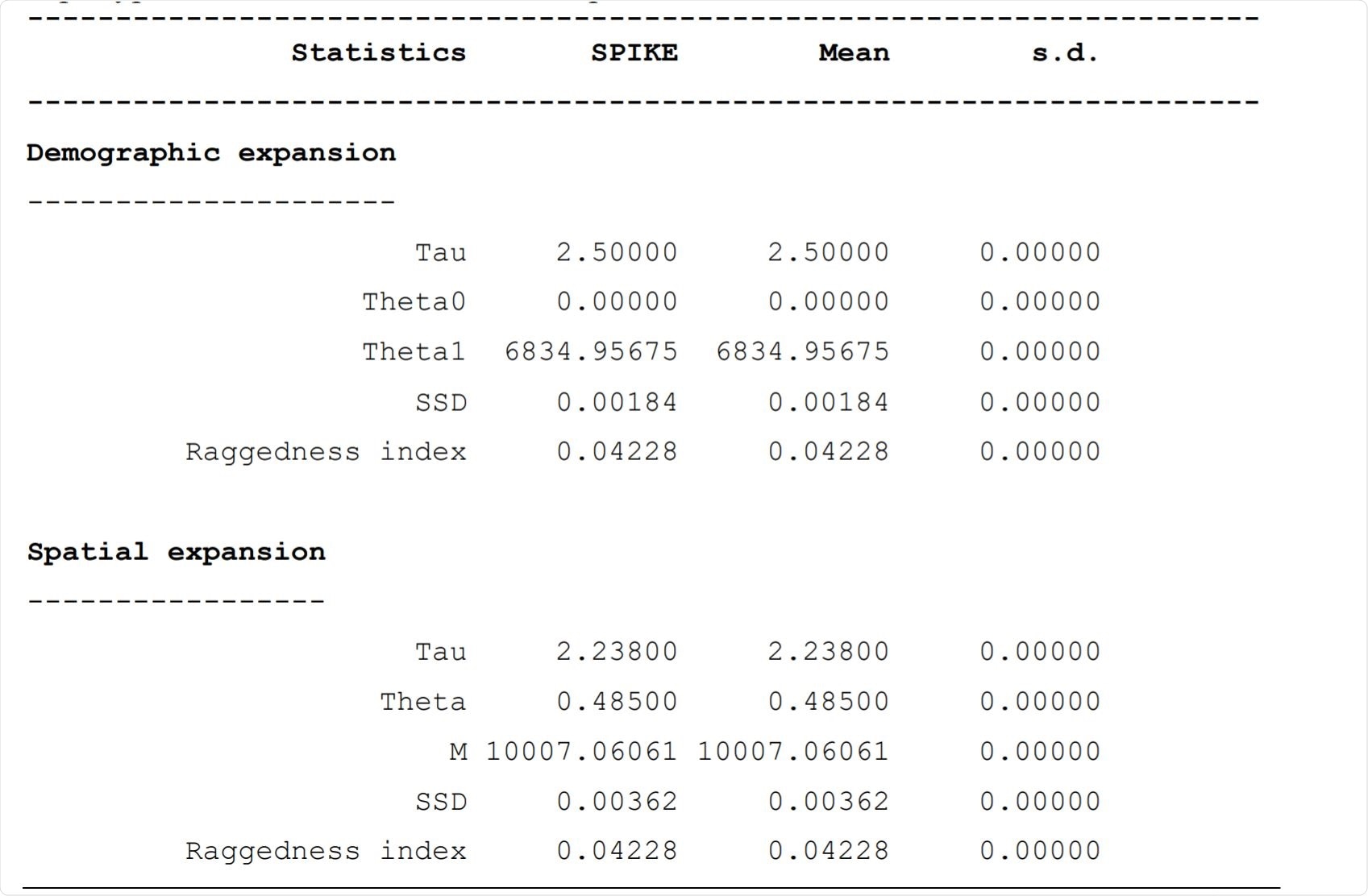
Mismatch analysis: demographic and spatial expansion rates for the 37 haplotypes of the SARS-CoV-2 SPIKE protein. Image Credit: https://www.biorxiv.org/content/10.1101/2020.12.16.423166v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The spike protein
The virus behind COVID-19 is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is known to mediate its entry into the host cell via its spike protein. This consists of a class I fusion protein, with one end embedded in the viral envelope and the other protruding from the surface. This protein both recognizes attaches to, and fuses with the host cell membrane to achieve internalization into the host cell.
The virus within the cell now proceeds to replicate its genome and to translate its structural and non-structural components, to assemble new full-grown virions. The exposed spike protein is a ready target for T cells, which are thereby activated to produce polyclonal antibodies to the several epitopes and specific domains of the spike protein. These antibodies are often neutralizing in nature, blocking virus attachment to the host cell receptor, the angiotensin-converting enzyme 2 (ACE2).
The spike protein is metastable in its prefusion form, which is composed of two subunits, S1 and S2. The first mediates binding to the receptor via its receptor-binding domain (RBD) and the second is responsible for virus-cell membrane fusion. This allows the virus to enter the cell, but this process is dependent on host cell proteases, which must cleave the subunits.
Spike target for antivirals and immunotherapies
The key role of the S protein has made it a prime target for immunotherapies and antivirals, as well as for vaccines under development.
Earlier, the discovery of the potent antigenic nature of the spike protein in SARS-CoV and MERS-CoV triggered the use of this protein as a vaccine antigen. In mouse studies, antisera collected from mice immunized with the S protein of the earlier SARS-CoV markedly reduced the virus entry into the target cells.
This suggests the induction of cross-reactive immunity against the spike protein epitopes, which are highly conserved in coronaviruses. Within 14 days post-immunization, both neutralizing antibodies and T cells targeting the spike protein are detectable. These animals were found to have low viral loads in the respiratory tract following subsequent exposure to the virus.
While other therapies are being explored, including extracts of medicinal plants and small molecules or peptides that target this protein, there is a long way to go, technologically speaking, before these become available as fully tested and approved commercially viable drugs.
Study findings
The current study aimed to explore the molecular diversity of the spike protein in different viral populations, which may have changed the genetic conformation and enabled immune evasion. The team of researchers, from the Laboratory of Population Genetics and Computational Evolutionary Biology (LaBECom-UNIVISA), carried out an analysis of molecular variance (AMOVA) on 37 different variants of the SARS-CoV-2 spike protein.
These haplotypes refer to linked sets of variations in the genome. All were retrieved from the publicly available National Biotechnology Information Center (NCBI) platform.
The researchers found a very low level of diversity between the haplotypes. The number of insertions, deletions, transitions, and transversions were very low. They observed only 17 polymorphic sites in the spike protein. The molecular diversity indices showed an absence of significant mutations for these haplotypes, and no significant population expansion was observed either.
What are the implications?
The researchers, therefore, found that SARS-CoV-2 spike haplotypes are highly similar. “We assume that molecular diversity for this protein, if found in future studies, may be associated with components of variation other than substitutions commonly found in the SARS-CoV-2 genome.”
These findings agree with the known conservation of the spike sequence. Thus, they say, the reported genetic variability of the virus spares some viral genes, leaving some ancestral proteins intact. The methods used in this study allowed for the discontinuous manner in which haplotypes developed genetic variations, via several mutational intermediate stages. The estimators used agreed with the uniformity of the results, despite their high sensitivity for any molecular variation.
These considerations ensure that the use of neutralizing antibodies may be able to suppress the proliferation of the virus, further justifying the development of vaccines based on protein S.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources