A team of scientists from the University of Saskatchewan, Canada, has recently demonstrated that chemical agents capable of reducing disulfide (S-S) bonds can be potentially used as antiviral drugs against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19). Their study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
As of January 5, 2021, over 84 million confirmed COVID-19 cases, including 1.8 million deaths reported to the World Health Organization (WHO). To reduce the transmission of SARS-CoV-2 and curb the COVID-19 pandemic trajectory, the global scientific community has been looking for therapeutic as well as preventive interventions. More than 180 vaccines candidates are being developed, and some of them have already shown affirmative responses in human clinical trials. Moreover, clinical studies conducted in search for effective therapeutic medicines have identified some repurposed antiviral medicines, such as remdesivir, hydroxychloroquine, lopinavir, and interferon, as potential agents to treat moderate-to-severe COVID-19 patients.
Mechanistically, the interaction between the spike protein (or S protein) of SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2) of the host cell is responsible for the viral entry. The structural analysis of SARS-CoV-2 has shown that the viral spike protein and its receptor-binding domain (RBD) contain multiple disulfide bonds (S-S), which are known to play vital roles in maintaining the structural and functional integrity of the spike protein. Moreover, there is evidence indicating that disruption of viral S-S bonds can lead to loss of its binding ability.
Given the abundance of S-S bonds in SARS-CoV-2 spike protein and their potential involvement in modulating the spike structure and function, the current study has been designed to evaluate the efficacy of disulfide-reducing agents in preventing SARS-CoV-2 infection in vitro.
Current study design
The scientists conducted molecular dynamics analysis and predicted that the reduction of S-S bond in the spike RBD makes the domain more flexible. Specifically, the analysis reveals that the S-S bond present in the surface loop of RBD (C480-C488) is responsible for maintaining the loop conformation, and thus, plays an important role in mediating the RBD – ACE2 interaction. As predicted by the molecular dynamics, the reduction of the S-S bond in the surface loop may affect the structural stabilization and impair the RBD – ACE2 interaction.
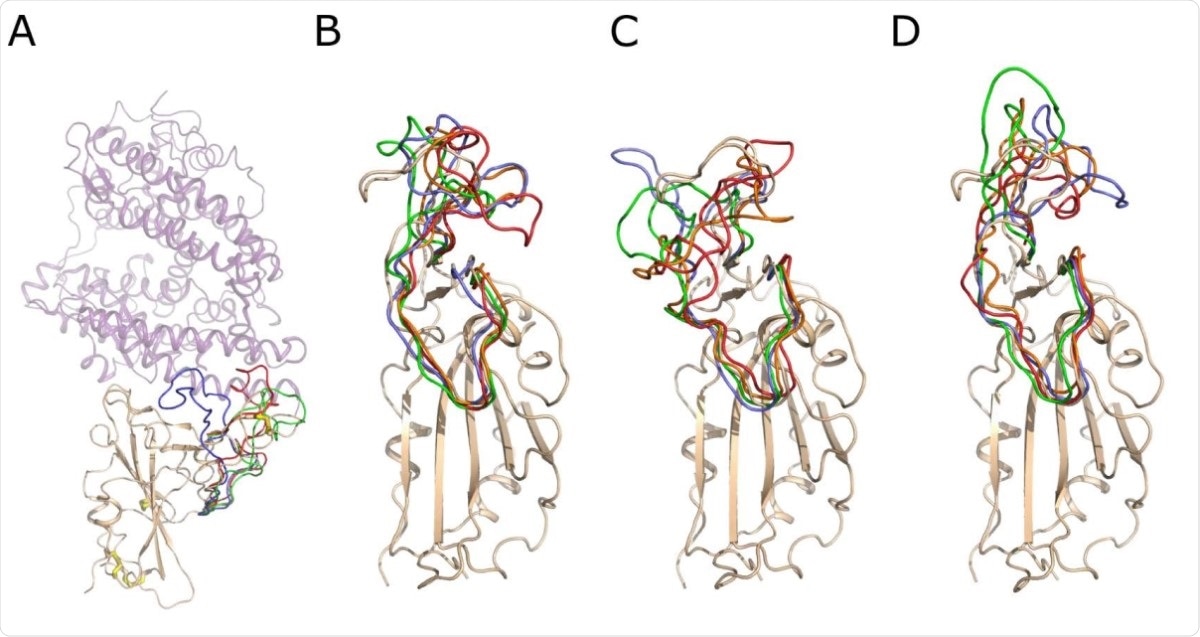
Snapshots of the conformation of the RBD surface loop, residues 456-490, along the molecular dynamics trajectories. A) The crystal structure of the Spike RBD – ACE2 complex. Spike RBD domain is shown as cartoon and colored wheat, while ACE2 is shown as a lilac semitransparent cartoon. The S–S bonds are shown as wire with white carbon and yellow sulfur atoms. The conformation of the RBD surface loop, residues 456-490, after 250 ns of molecular modeling is shown in green, red and lavender for RBD, RBD (-4 SS) and RBD (-1 SS), respectively. B) Snapshots of conformations of the RBD loop 456-490. The RBD domain is shown as a wheat cartoon. The structure of the loop is shown in lavender, green, orange and red color for 250, 500, 750 and 1000 ns of the modeling trajectory. C) Snapshots of the loop 456-490 conformations of the RBD (- 1 SS). D) Snapshots of the loop 456-490 conformations of the RBD (-4 SS). The coloring scheme is identical for B, C and D. The structural deviations of the loop 456-490 are much higher if the Cys480-Cys488 bond is reduced, as shown in C and D.
Based on the molecular dynamics predictions, the scientists have conducted a series of in vitro and cell-based assays to determine the ability of several disulfide-reducing agents, such as dithiothreitol (DTT), tris(2-carboxyethyl)phosphine (TCEP), N-Acetyl-L-cysteine (NAC), and reduced glutathione (GSH), in mitigating SARS-CoV-2 infection.
Important observations
The data obtained from in vitro assays reveal that disulfide-reducing agents DTT and TCEP are capable of altering the secondary structure composition of the spike RBD and reducing its melting temperature almost to the physiological range. Moreover, these two chemical agents significantly impair the interaction between RBD and ACE2 by reducing the RBD S-S bond. A very low melting temperature of RBD observed in the presence of DTT and TCEP indicates that administration of reducing agents into the human body can lead to an unfolding of RBD even at normal body temperature. Other reducing agents tested in this study, such as NAC and GSH, demonstrate similar effects, but the impact is lesser than DTT and TCEP. In contrast to RBD effects, incubation of DTT and TCEP with ACE2 does not cause any S-S reduction, indicating that ACE2 is resistant to external reducing agents.
Regarding the interaction between RBD and ACE2, both DTT and TCEP significantly reduce the binding affinity, whereas NAC and GSH show a less significant impact.
In line with in vitro experiments, the data obtained from cell-based analyses demonstrate that both DTT and TCEP can prevent SARS-CoV-2 entry and replication inside human cells. The concentrations of DTT and TCEP used to achieve the antiviral effects have not caused any reduction in cell viability, indicating that these agents can be used therapeutically to prevent SARS-CoV-2 transmission.
Of four tested reducing agents, DTT and TCEP demonstrate the highest effects as they have higher redox potential than NAC and GSH, which are mono-thiols and require the presence of other thiol-containing molecules to catalyze S-S reduction.
Overall, the study findings provide useful information about the potential antiviral effects of disulfide-reducing agents. Given their ability to inhibit the viral entry into host cells, these reducing agents may be used as the first line of defense against SARS-CoV-2 infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
BioRxiv preprint server. 2021. Grishin AM. Spike protein disulfide disruption as a potential treatment for SARS-CoV-2. doi: https://doi.org/10.1101/2021.01.02.425099, https://www.biorxiv.org/content/10.1101/2021.01.02.425099v1
- Peer reviewed and published scientific report.
Grishin, Andrey M., Nataliya V. Dolgova, Shelby Landreth, Olivier Fisette, Ingrid J. Pickering, Graham N. George, Darryl Falzarano, and Miroslaw Cygler. 2021. “Disulfide Bonds Play a Critical Role in the Structure and Function of the Receptor-Binding Domain of the SARS-CoV-2 Spike Antigen.” Journal of Molecular Biology, November, 167357. https://doi.org/10.1016/j.jmb.2021.167357. https://www.sciencedirect.com/science/article/pii/S0022283621005945.