Over the past year, there was a notable evolution of SARS-CoV-2 around the globe, which will continue to pose a serious threat. As of January 2021, four novel viral variants have been reported from the United Kingdom (VOC 202012/01/B.1.1.7), South Africa (501Y.V2), Nigeria (B.1.207), and Denmark (Mink Cluster V).
Such a fast and constant pace of evolutionary changes affects the pathogenicity of SARS-CoV-2, a causative agent of coronavirus disease (COVID-19), and definitely warrants additional studies. In other words, an in-depth look at the genome is necessary to appraise all genomic alterations and adaptations.
It is already known that mutations in the viral spike glycoprotein can modify binding efficiency and, in turn, viral fitness. In fact, certain nucleotide mutations in the S gene of the SARS-CoV-2 can change its pathogenicity and diminish virulence, which is the reason why they have become pervasive in tracking the spread of the virus.
In this study, the researchers from the University of Hawaii (led by David P. Maison, MS) report the analysis of a 969-bp SARS-CoV-2 S gene from two patients from Hawaii in order to appreciate changes in the spike glycoprotein, which is also a prime target used by vaccines.
Generating phylogenetic trees
At the John A. Burns School of Medicine Biocontainment Facility of the University of Hawaii, ribonucleic acid (RNA) was extracted from a nasal swab and an oropharyngeal swab from two male patients (mean age of 29.5 years) infected with the SARS-CoV-2 in August 2020.
After pursuing polymerase chain reaction (PCR), the two viral strains were sequenced with the use of Sanger sequencing (also known as the "chain termination method"). The latter was conducted on the amplicons by utilizing four primers (CF, CR, CR2, and CR3).
Furthermore, phylogenetic trees were generated using MEGAX (i.e., an integrated tool for automatic and manual sequence alignment). The alignment was initially done by using the program MUSCLE, and then it was generated with Maximum Likelihood parameters with 1,000 bootstraps.
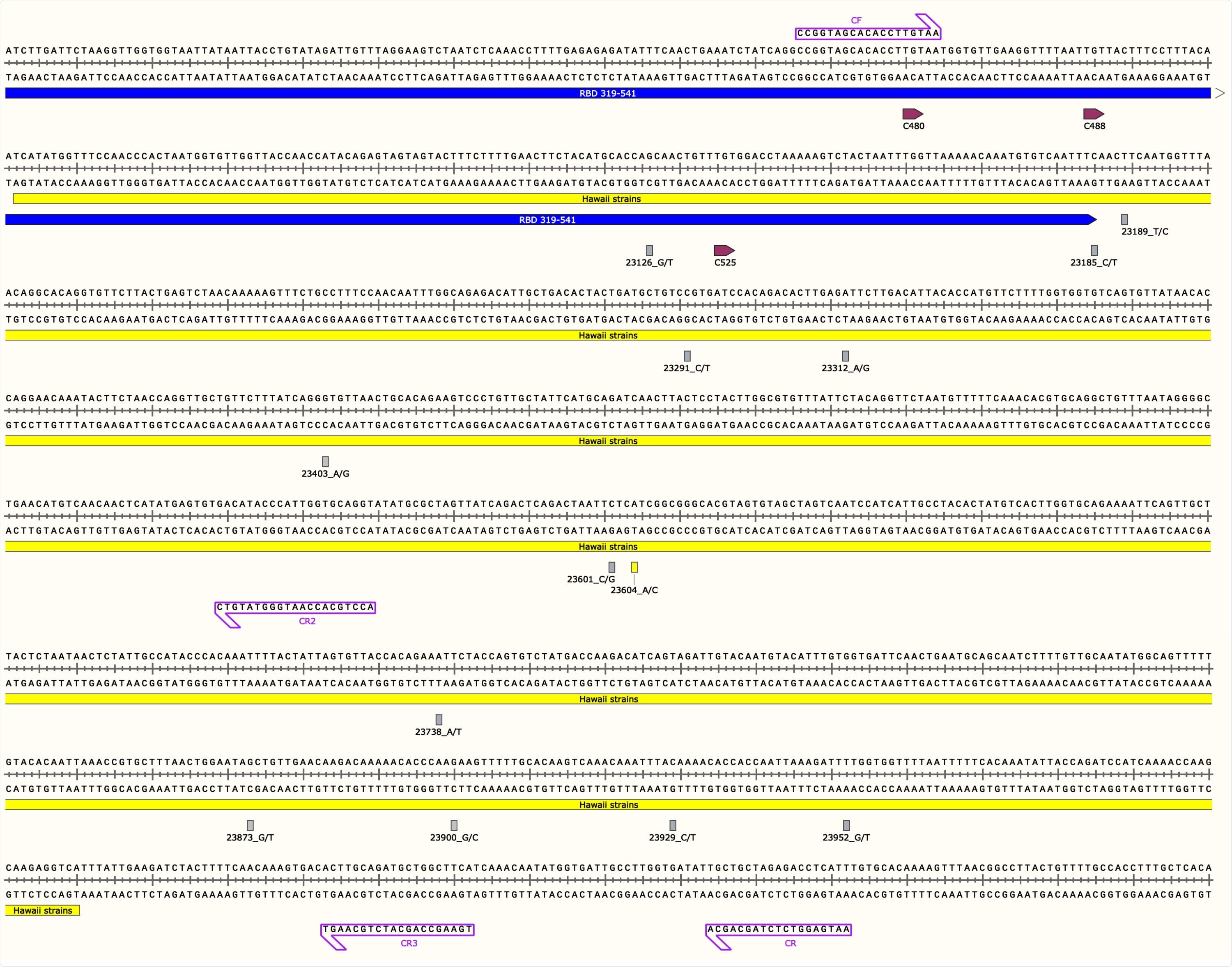
SARS-CoV-2 S Gene Region used in this Study Along with Annotated Primers, Mutations, and Cysteine Residues of the Receptor Binding Domain Figure represents the Hawaii strain MW237663 and MW237664 sequences. The primer pair, CF/CR, was used to amplify the 1,127-bp S gene fragment and primers CF, CR, CR2 and CR3 depicted with purple boxes were used for Sanger sequencing. The yellow box indicates the start and end of the 969-bp sequence. The blue line indicates the 3’ end of the S gene receptor binding domain (RBD). RBD cysteine residues are shown in depicted boxes. All mutations found in this study are in their respective loci with nucleotide numbers and rectangular boxes correlating to the sense strand as indicated after the nucleotide number and underscore in the figure (nucleotide/protein mutations: G23126T/A522S, C23185T/F541F, T23189C/F543L, C23291T/R577C, A23312G/I584V, A23403G/D614G, C23601G/S680C, C23604A/P681H, A23738T/I726F, G23873T/A771S, G23900C/E780Q, C23929T/Y790Y, and T23952G/F797C). All boxes are grey except for the P681H mutation seen in the Hawaii strains from this study, shown with a yellow rectangle. Image was generated with the SnapGene software (from Insightful Science; available at snapgene.com) and was created with BioRender.com.
Emerging traits of P681H mutation
In short, Hawaiian SARS-CoV-2 strains that were deposited in the GenBank in March 2020 clustered with sequences from Wuhan, China, Sweden, and the state of New York (USA). The SARS-CoV-2 strains in this study clustered from the state of Washington (USA), as well as with sequences from China and Hawaii.
Moreover, phylogenetic tree results suggest that the virus has been brought to Hawaii from many sources. Thirteen single nucleotide polymorphisms were decoded across 13 unique SARS-CoV-2 genomes within the S gene region – with one non-synonymous mutation (P681H) detected in the two Hawaii strains.
This notorious P681H mutation has distinctive and emerging characteristics, with an exponential increase in global frequency in comparison to the plateauing of the (currently universal) D614G mutation. Likewise, the P681H mutation is also a characteristic feature of the new SARS-CoV-2 variants found in the United Kingdom and Nigeria.
Several mutations resulting in cysteine residues were also found, with potential consequences such as the disruption of the disulfide bridges in and around the receptor-binding domain of the viral spike glycoprotein.
A potential viral evasion mechanism
This study actually demonstrates a partial sequence from the first SARS-CoV-2 strain that harbors the non-synonymous P681H mutation. Further studies are, therefore, warranted to analyze the pathogenicity and virulence of this mutation in the Hawaii strains and whether it can be considered a viral evasion mechanism to put off antibody recognition.
"In Hawaii, Native Hawaiians and Pacific Islanders have a significantly high prevalence of SARS-CoV-2 when compared to other ethnic minorities and Whites", say study authors in this bioRxiv paper. "Characterizing viral sequences from these minority groups is important to better understand virus transmission and pathogenicity", they add.
Going forward, targeted sequence characterization will be necessary to determine the origin of multiple introductions of SARS-CoV-2 circulating in Hawaii, which can subsequently inform similar studies in other parts of the world.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.