The resurgence of the coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to renewed efforts to find effective antivirals, both newly developed or repurposed drugs. A new preprint appearing on the bioRxiv* server reports the efficacy of the cholesterol-reducing drug fenofibrate in reducing infection rates of SARS-CoV-2 in cultured cells.
The virus must engage host cells via its receptor-binding domain (RBD) on its spike protein, which binds to the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell. Inhibitors of spike-ACE2 binding are therefore likely to reduce the entry of the virus into host cells, thus reducing disease severity or preventing transmission of the virus altogether.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Among the drugs sought to be repurposed for use against SARS-CoV-2 are dexamethasone and remdesivir. Since these drugs are already available for use, and have well-characterized pharmacological profiles, they can be rapidly introduced into clinical use once their efficacy is established.
Study details
The current study employed an assay that evaluates the spike RBD-ACE2 binding to help identify binding inhibitors that act through novel mechanisms, and might therefore escape recognition. ACE2 is a dimer, and multiple RBDs may interact with each ACE2 molecule. It is also a flexible protein, enabling binding between more than one dimeric ACE2 with a single trimeric spike. If ACE2 dimerization is affected, therefore, the avidity of binding to RBD is possibly impaired.
With other receptors, dimer formation has been found to increase internalization. This could therefore be a promising target for antiviral drugs. The researchers developed another assay that measures dimerization of ACE2 using the NanoBIT interaction system.
The system contains two components, LgBIT and SmBIT, which are individually inactive but are activated when they combine to form an active luciferase enzyme. Thus, they can be fused to the ACE2 monomer so that their association in dimer form allows the formation of active luciferase.
Fenofibrate promotes ACE2 dimerization
This was then used to look through a library of a hundred already approved drugs to identify potential inhibitors of ACE2 dimerization. They found that fenofibric acid, which is the active form of the cholesterol synthesis inhibitor fenofibrate, led to twice as much ACE2 dimer formation, which prevented RBD-ACE2 binding by destabilizing the spike RBD. Both fenofibrate and fenofibric acid reduced the thermal stability of RBD, though fenofibric acid was active at a much lower drug concentration. The fenofibric acid had a modest but significant inhibitory effect on RBD-ACE2 binding, as did fenofibrate.
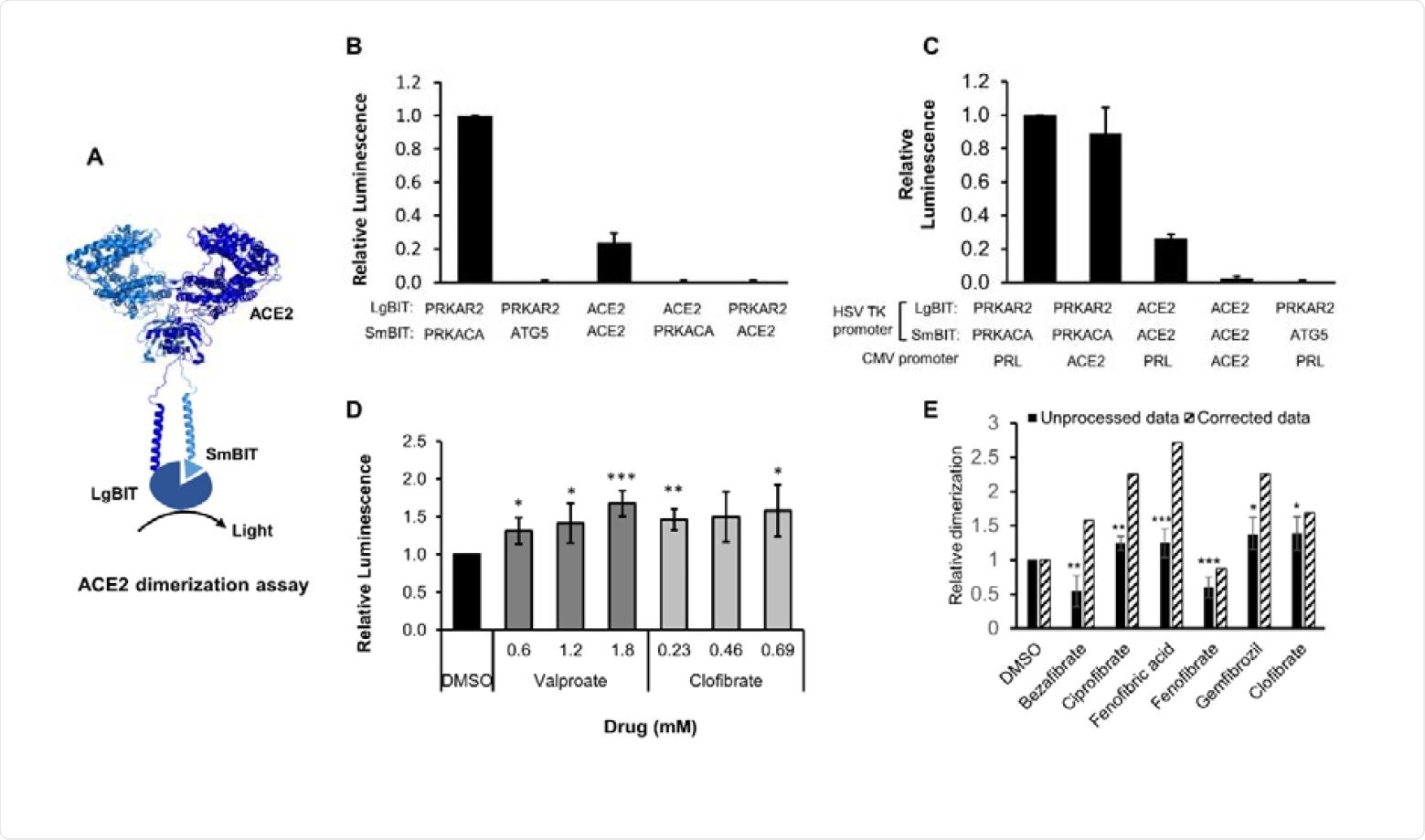
ACE2 dimerization assay. A. Schematic showing ACE2 tagged with LgBIT and SmBIT. B. HEK-293 cells were transfected with combinations of plasmids encoding LgBIT or SmBIT fused to either protein kinase A regulatory subunit (PRKAR2) or catalytic subunit (PRKACA), ATG5 or ACE2. The results (mean ± S.D., n = 5) were normalized to the luminescence measured in cells transfected with protein kinase A reporters (positive control). C. HEK-293 cells were transfected with plasmids encoding ACE2 nanoBIT reporters under the control of the HSV TK promoter and ACE2 or prolactin (PRL) under the control of the CMV promoter. The results (mean ± S.D., n = 4) were normalized to the luminescence measured in cells transfected with protein kinase A reporters and prolactin. D. HEK-293 cells were transfected with NanoBIT-tagged ACE2 reporters and incubated with sodium valproate or clofibrate at a concentration equal to 1x, 2x or 3x the reported Cmax of the drug. After 1 hour, luminescence was measured and normalized (mean ± S.D., n = 4) to that measured in cells treated with DMSO. E. A series of other fibrates were similarly evaluated in the assay. The luminescence measured (mean ± S.D., n = 5-11, solid bars) was significantly different to that measured in cells treated with solvent where shown. When these fibrates were incubated with purified LgBIT and HiBIT-RBD to create a constitutively active nanoluc, each of these fibrates were found to inhibit nanoluciferase (bezafibrate 35 ± 7 %, ciprofibrate 55 ± 6 %, fenofibric acid 46 ± 3 %, fenofibrate 69 ± 5 %, gemfibrozil 61 ± 2 % of the activity measured in the presence of DMSO). To correct for this, the luminescence measurements from cells treated with fibrates in cells were divided by these latter values to estimate the effect of the drugs on dimerization (hatched bars). Significant difference from control is shown as *, P< 0.05; **, P < 0.01; ***, P< 0.005.
The increase in dimerization peaked at about 30 minutes following exposure to fenofibric acid. Fenofibrate failed to show any significant activity in this assay, perhaps because it lacks a free carboxylic acid, unlike fenofibric acid.
Inhibition of infection in cell culture
The next step was to screen for the effect of the drug on the virus infection itself.
The two drugs were assessed for infection inhibition by SARS-CoV-2 in two different assays and using two different viral isolates. The use of live virus allowed both primary infection and secondary infection by newly released viral particles to be assessed, at 24 and 48 hours, respectively.
In the first step, the hCOV-19/England/2/2020 virus strain was tested. With fenofibrate, virus infection was reduced by 65% (from 59% of cells to 18% of cells) at 230 μM, relative to the controls.
Cell viability was not reduced. In contrast, other statins like simvastatin and pitavastatin also reduced infection rates but at the cost of cell viability.
Secondly, they tested the effect of fenofibric acid and fenofibrate using live SARS-CoV-2. They found that at 24 hours, fenofibrate reduced infection levels by 60%, and to a smaller extent with fenofibrate, though the reduction observed with fenofibric acid was not significant. The same pattern was seen at 48 hours, which showed sustained suppression of infection by fenofibrate.
This agrees with earlier studies showing that fenofibrate, as well as fenofibric acid to a smaller extent, are strong contenders as SARS-CoV-2 inhibitors. The pattern of inhibition was sustained at 24 and 48 hours. Thus, fenofibrate can both prevent primary infection and reduce secondary infection rates.
This was confirmed by reduced viral N-gene RNA levels with fenofibrate, in a dose-dependent manner. Even though fenofibrate operates on lipid metabolism via the PPARα pathway, this does not appear to account for the antiviral action.
Inhibition of plaque formation
The result was confirmed using a second strain of the virus in a plaque assay. The use of fenofibric acid reduced plaque formation by 76% at a dose of 250μM, with fenofibrate showing a similar but less powerful action. Since plaque assays are the gold standard in determining infectivity for this SARS-CoV-2, these results are significant. In both cases, pretreatment with fenofibrate was just as powerful as co-treatment.
Possible mechanisms of action
It appears that the antiviral effects of fenofibrate and its metabolite are not due to its inhibition of cholesterol synthesis, nor to their direct effect on the viability of the infected cells. The differences in the level of inhibition observed with the different assays may be due to the different strains used or the variation in the methods. For instance, fenofibrate may alter the conformation of ACE2 rather than causing it to dimerize.
The ‘open’ and ‘closed’ conformations of ACE2 may affect RBD-ACE2 binding at the level of RBD monomers, or affect the number of spike proteins capable of attaching to each ACE2 dimer. Either way, binding avidity could be affected. Changes in ACE2 conformation could also make it more resistant to proteolysis by the serine protease TMPRSS2.
If fenofibrate truly does exert its antiviral activity via ACE2-mediated effects, this imparts additional advantages over other drugs that inhibit viral proteins, because its activity is less susceptible to viral mutational escape. Secondly, it reduces the RBD stability and impairs ACE2-RBD binding, both of which reduce the extent of infection in treated cells.
This again shows that fenofibrate may act via multiple mechanisms, reducing the chances of viral resistance emerging rapidly. For instance, it increases the levels of the glycosphingolipid sulfatide, which could reduce SARS-CoV-2 infection. It suppresses airway inflammation and hence cytokine release, which is associated with severe COVID-19. It has also been suggested to reduce thrombosis and platelet aggregation, promote clot lysis and thus prevent or modulate hypercoagulability, seen in advanced COVID-19. Its role in inhibiting cholesterol biosynthesis via PPARα has also been suggested to be beneficial in preventing infection, and two trials have been registered to examine this premise.
What are the implications?
The study concludes, “Using two different virus isolates, we demonstrate that fenofibrate, or its active metabolite fenofibric acid, are able to significantly reduce SARS-CoV-2 infection in cell culture models.”
The safety of this drug, its convenience of use, and bioavailability at currently recommended doses, along with its cheapness and wide availability as a generic drug, has led to the recommendation that it be used in clinical trials to assess its efficacy in preventing severe or critical COVID-19.
Further studies to clarify the precise mechanism of the anti-viral activity of fenofibrate are desirable, but this should not delay the urgent clinical evaluation of the drug to counter the current pandemic.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources