Using more than 100,000 virus genome sequences uploaded in the GISAID database, researchers found around 9,000 mutations in the virus spike protein RBD.
The virus's spike protein interacts with host receptors like the angiotensin-converting enzyme 2 (ACE2) and protease TMPRSS2, which allow the virus to bind and then fuse with the host cell. The virus releases RNA into the host cytoplasm, where it replicates.
The spike protein has two subunits, S1 containing the receptor-binding domain (RBD), which interacts with ACE2, and S2. Mutations in the RBD or ACE2 can affect virus infectivity and disease condition.
To understand RBD mutations and their effect on antibody interactions, a team of researchers from the University of Toronto analyzed the virus RBD using the SARS-CoV-2 genome information in the Global Initiative on Sharing All Influenza Data (GISAID) database. They report their results in a research paper posted to the bioRxiv* preprint server.
RBD Mutations
The researchers obtained more than 110,000 RBD sequences uploaded on the database until 15 October 2020. After removing duplicates, sequences with gaps, and other irrelevant sequences, the team obtained 106,941 sequences. The nucleotide sequences were aligned to the RBD of the reference strain and analyzed.
Compared to the Wuhan reference strain, the team found 9,275 mutations on the RBD, leading to 9,064 mutant SARS-CoV-2 strains. About 75% of the nucleotide mutations were G1430A, 6.8% were C1317A, and 0.9% were G1144T. The most common mutation was a change from G to A, occurring in about 7% of the mutants, followed by changes from C to A, then G to T, and C to U.
Of all the nucleotide mutations, about 98.3% changed the amino acid sequence of the RBD. The rest were synonymous mutations. The G1430A change led to the S477N mutant, representing 76.5% of all the amino acid mutations. The C1317A and the G1144T mutations led to the N439K and A520S mutations, respectively.
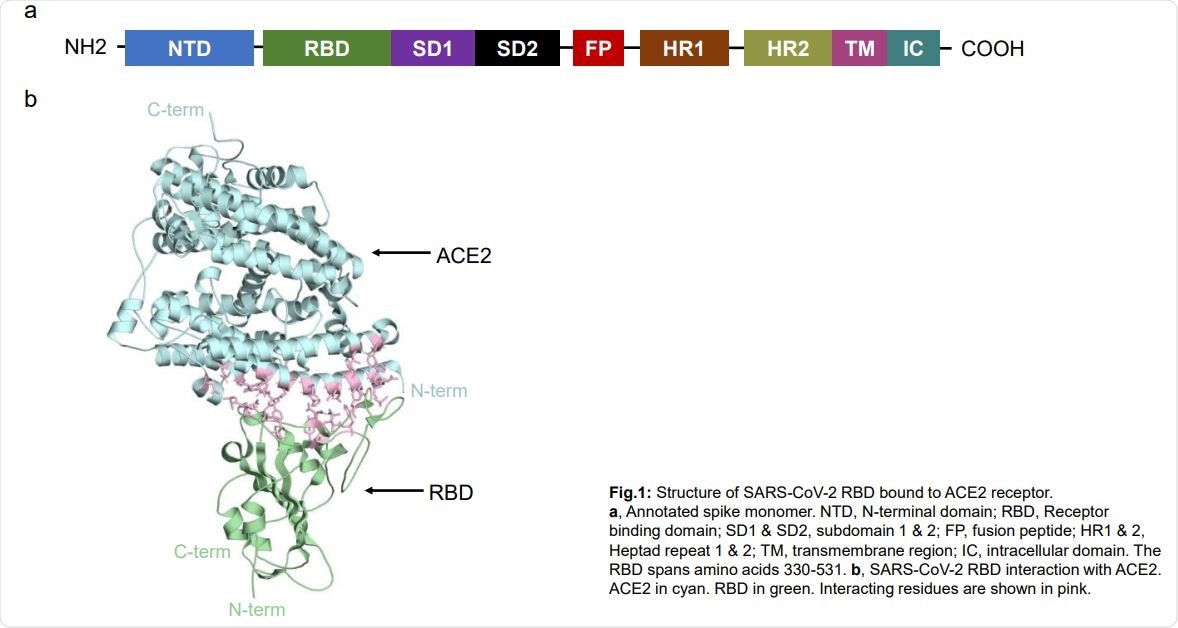
Structure of SARS-CoV-2 RBD bound to ACE2 receptor. a, Annotated spike monomer. NTD, N-terminal domain; RBD, Receptor binding domain; SD1 & SD2, subdomain 1 & 2; FP, fusion peptide; HR1 & 2, Heptad repeat 1 & 2; TM, transmembrane region; IC, intracellular domain. The RBD spans amino acids 330-531. b, SARS-CoV-2 RBD interaction with ACE2. ACE2 in cyan. RBD in green. Interacting residues are shown in pink.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Previous studies have shown that the S477N and N439K mutants show increased affinity to ACE2. The analysis showed these mutations represent 6.7% and 0.6% of the total genomes in the database, respectively, with a 70-fold increase in the presence of the S477N mutation. This suggests selection could be playing a role in the RBD mutations. In total, the authors found 13 mutations leading to an increase in the binding affinity with ACE2.
The S477N mutation was found in 14 countries worldwide, with about 69% of Australia's sequences having this mutation. N439K was seen in 10 countries, with the highest occurrence in England. They did not find both mutations occurring on the same genome. The team also found that all the S477N or N439K mutations also had the D614G mutation in the spike protein.
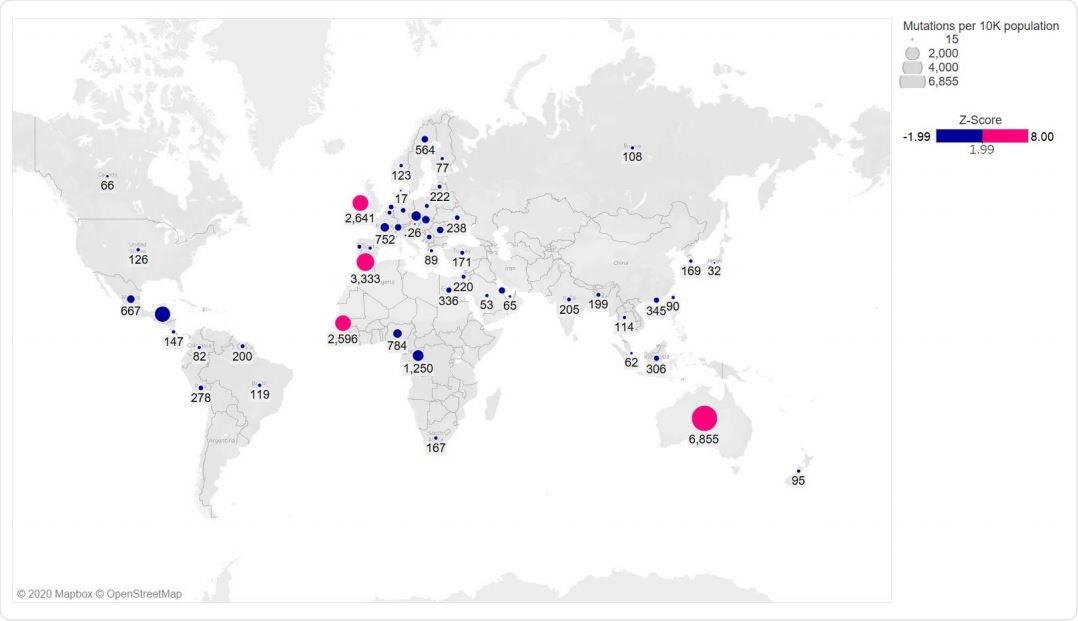
Global distribution of RBD mutations
No mutations at residue that increases binding affinity to ACE2
What surprised the authors was that they did not find any mutations at the Q498 residue of the RBD because Q498H, Q498Y, and Q498F all increase binding affinity to ACE2. They also either did not find or found in very now numbers other mutations, such as N501F, Y453F, T385R, Q493M, and Q414A, which increase RBD binding to ACE2.
This may be because mutations at Q498 would be disadvantageous to virus fitness and might reduce virus transmissibility according to the transmission-mortality trade-off theory. If this were to be accurate, the mortality rate of SARS-CoV-2 would decrease over time, and transmission will increase.
When the team analyzed human antibody interaction sites with the RBD, they found 75% of the interaction sites of 10 human antibodies had mutations. Their analysis also showed that the antibodies worked by broad coverage of the RBD. None of the mutations had any direct interaction with every antibody they studied.
“This suggests that immunization with wild type and potentially any RBD point mutant that conserves structure will likely elicit the development of RBD antibodies sufficient for binding and resolving infection,” write the authors. However, it remains to be seen how the mutations will affect virus infectivity and transmissibility.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources