The coronavirus disease 2019 (COVID-19) pandemic has now caused over 100 million cases, with more than 2 million deaths worldwide. Developed nations continue to grapple with increasing caseloads caused by infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19. In the USA alone, thousands of deaths continue to be reported daily.
This has made it a vital necessity to develop a test enabling the early detection of infection before it produces symptoms, thus allowing for transmission chains to be broken.
An encouraging new study by researchers at University College London, UK, reports the potential of a biomarker test that measures the levels of IFI27 in blood, and could perhaps allow this goal to be reached.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The research team has published their findings on the medRxiv* preprint server.
Issues with current diagnostic tests
It has been established that viral infectivity is maximal during the first week following infection. This period should therefore be the focus of case detection efforts in order to effectively isolate infected individuals, along with contact tracing and quarantine. Screening tests to identify infection in the asymptomatic and mildly symptomatic cases are important tools to achieve this level of containment, especially for mass testing.
Available tests today include lateral flow assays (LFAs) that detect viral antigens, but these are not sensitive enough to rule out active infection, thus making them unsuitable for contact screening or mass screening. The gold standard is the reverse-transcriptase PCR (RT-PCR) test, which identifies viral RNA. This is, however, both expensive and time-consuming, and requires skilled equipment and operators. The time required for a PCR test may be reduced by LAMP (loop-mediated isothermal amplification) assays which are, however, less sensitive. Moreover, all these tests require swabs from the nasopharynx or throat, which requires operator skill. And finally, the high false-negative rates of all these tests limits their use for mass screening.
Blood tests have many advantages, in contrast to the above tests. The current study aims to evaluate the transcription markers of viral infection in blood in order to possibly develop a candidate for prediction of PCR positivity for SARS-CoV-2 in healthcare workers (HCWs) who are tested every week.
Study details
The study included 96 participants, who gave 169 blood samples for RNA analysis.
The study showed that Interferon Alpha Inducible Protein 27 (IFI27) was the most specific discriminatory parameter for SARS-CoV-2 infection, as confirmed by PCR. The sensitivity as determined by a value two standard deviations above the value in healthy controls was 84%, and the specificity 95%. This compared favorably with three other transcriptional signatures, of which IFI27 was lacking in only one. The scores for the highest performing signatures were negatively associated with PCR cycle thresholds (Ct), suggesting that as viral loads increased, these signature scores also went up.
The maximum scores were in the first week following the initial PCR positivity result and were normal in convalescent serum. Scores in the week before the first positive PCR were also higher than in either controls or convalescent serum. The discriminatory values for scores at this time were also statistically valid but lower than for scores at the time when PCR was positive. By way of example, one week preceding the first positive score, the IFI27 was able to predict infection with 79% accuracy, with 40% sensitivity and 95% specificity.
The researchers say this is the first time host transcription profiles have been assessed for their use in the detection of presymptomatic SARS-CoV-2 infection. It capitalizes on the accuracy of the IFI27 gene that is the best-performing marker in a set of signatures found to have high predictive value for this infection relative to controls.
The connection of these signatures with type I interferon responses is significant, since the latter is a specific feature of antiviral responses in the host. Severe COVID-19 is more common when this pathway is affected by genetic or immune system dysregulation factors. IFI27 has been studied for its role in type I IFN-dependent apoptosis, as part of interferon effects on tumor growth. More work may show how such IFN-inducible genes are regulated during this infection.
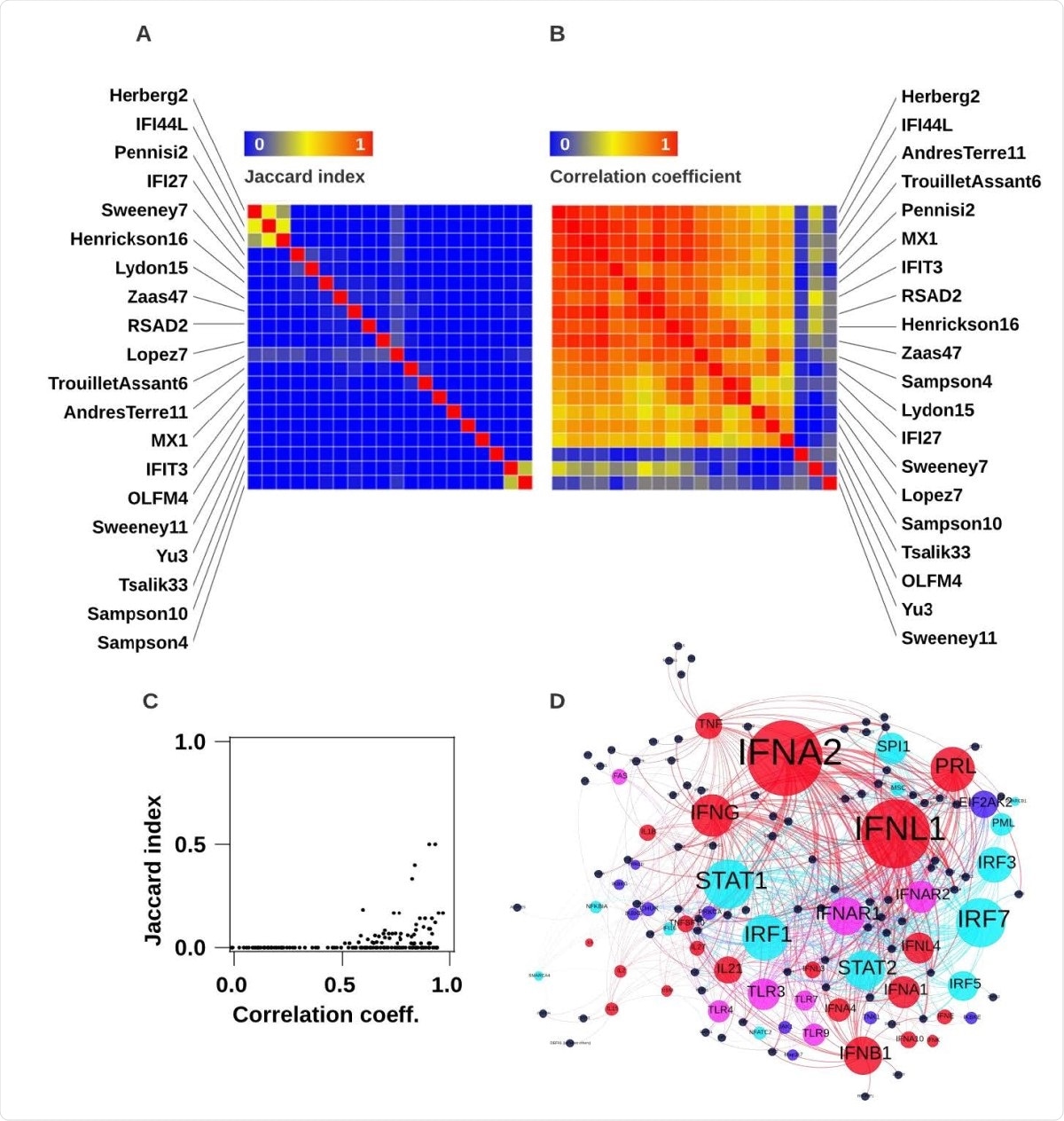
Correlation and Jaccard indices for all RNA signatures for viral infection included in analysis. (A) Jaccard index intersect of constituent genes for all pairs of signatures clustered by Euclidean distance. (B) Spearman rank correlation coefficients for all pairs of signatures clustered by 1-Spearmean rank distance (C) relationship between pairwise Jaccard indices and Spearman rank correlation coefficients. (D) Network plot of significantly enriched predicted upstream regulators by cytokine (red nodes), transmembrane receptors (purple nodes), kinase (dark blue nodes) and transcription factors (light blue nodes) of all constituent genes in any signature (black nodes). Size of upstream regulator nodes proportional to statistical enrichment (-log10 FDR).
What are the implications?
Our data therefore demonstrate that measurable type 1 IFN-stimulated 134 responses to SARS-CoV-2 precede the onset of symptoms, and in some individuals may predate detectable viral 135 RNA on RT-PCR testing.”
This could be the basis of new tests to diagnose the infection in the presymptomatic stage by the detection of transcribed viral RNA signatures, which not only provide a means for very early diagnosis but also associate with viral loads. This may make it possible to identify the individuals who are most infectious, allowing transmission to be interrupted.
The study applies at present only to presymptomatic infection and may not be useful in severe or critical COVID-19. They are found in multiple viral infections and could thus be best viewed as a potential tool for screening contacts to allow infection control and identify those who might most probably be PCR positive.
If translated to a near-patient diagnostic test, this transcript could have significant clinical utility by facilitating early case detection.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Gupta, R. K. et al. (2021). Blood transcriptional biomarkers of acute viral infection for detection of pre-symptomatic SARS-CoV-2 infection. medRxiv preprint. doi: https://doi.org/10.1101/2021.01.18.21250044. https://www.medrxiv.org/content/10.1101/2021.01.18.21250044v1
- Peer reviewed and published scientific report.
Gupta, Rishi K, Joshua Rosenheim, Lucy C Bell, Aneesh Chandran, Jose A Guerra-Assuncao, Gabriele Pollara, Matthew Whelan, et al. 2021. “Blood Transcriptional Biomarkers of Acute Viral Infection for Detection of Pre-Symptomatic SARS-CoV-2 Infection: A Nested, Case-Control Diagnostic Accuracy Study.” The Lancet Microbe 2 (10): e508–17. https://doi.org/10.1016/s2666-5247(21)00146-4. https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(21)00146-4/fulltext.