The coronavirus disease 2019 (COVID-19) pandemic has caused huge devastation to human life and wellbeing. The present difficulties are unlikely to end unless population immunity is achieved. The causative agent of COVID-19, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has spread to every region of the world, with over 106 million documented infections and more than 2.3 million casualties so far.
A team of researchers recently examined the SARS-CoV-2 transcriptome and the dynamics of the S gene furin cleavage site in primary human airway epithelia. The team has released their findings on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
SARS-CoV-2 spike
This virus has a large 30 kb genome, encoding its structural and non-structural proteins, as well as several accessory proteins. The spike proteins of the various coronaviruses share 77% homology.
The SARS-CoV-2 spike protein has two subunits, the S1 and S2. S1 mediates viral spike-host cell angiotensin-converting enzyme 2 (ACE2) receptor binding, while S2 mediates viral entry into the host cell via membrane fusion.
The spike protein is the immunodominant antigen in human infection, as well as playing the central role in the type of host cell targeted by the virus, the pathogenicity of the virus, and its transmissibility. In nature, it occurs as a homotrimer on the surface of the viral particle.
Importantly, the SARS-CoV-2 spike is different from that of the closely related bat SARS-like coronaviruses encoding a unique polybasic furin cleavage site (FCS) at the S1/S2 interface. This is composed of four amino acids, PRRA.
The FCS gene itself is composed of the RRAR↓S sequence. Its presence in this virus, but not in other betacoronaviruses, indicates that it is crucial to the virulence of the virus by increasing the host range, the range of cell tropism, its transmissibility, and pathogenicity.
Rapid emergence of FCS deletions
When grown in Vero cells, derived from African green monkey cells, SARS-CoV-2 rapidly produced S1/S2 interfacial mutations that included the loss of the FCS with the proximal and distal amino acids in its progeny virions. These formed a stable dominant population with repeated passaging.
The spike gene is the most variable among all SARS-CoV-2 genes, and the FCS region at the S1/S2 interface is especially prone to mutations. A variety of deletions and mutations have been identified in this region in Vero cells infected with the virus.
Especially, one mutant comprising a 30 base pair deletion was able to replicate more efficiently within Vero cells, becoming the dominant strain on passaging. Another 21-base pair deletion was also found after two or three passages. These have not been found in the clinical isolate that was originally passaged, however. This shows that these were acquired during Vero cell passages.
Clinical isolates show the presence of many deletions around the FCS. The current study aimed to understand how these arose during the growth of the virus in human airways.
Study details
The researchers used RNA sequencing methods to explore the virus's transcription in infected human airway epithelium (HAE) cells, cultured to simulate natural infection of the human airways. This was achieved by culturing them at an air-liquid interface (HAE-ALI).
This method of culture yielded a transcriptomic pattern that more closely resembled that in ordinary human airway epithelium in COVID-19 patients. In both HAE-ALI and Vero cell cultures infected with SARS-CoV-2, nucleocapsid (N) transcripts, in the form of subgenomic RNA (sgRNA), were abundant, with lower levels of spike (S) transcripts.
They found that significant differences involved chiefly the deletions at the S1/S2 interface. The high rate of FCS deletions contributed to a much lower percentage of sgRNAs relative to all canonical sgRNAs, at only 50%, in infected HAE-ALI cells.
Two deletions in the FCS were found at dramatically enhanced levels in two cultures. One is a 12 amino acid deletion (mut-del1), 678TNSPRRAR↓SVAS689, including the FCS. The second is a deletion of 5 amino acids (mut-del2), 675QTQTN679, immediately upstream of the FCS.
Donor-specific selection
They observed donor-specific selection pressure operating within these HAE-ALI cells to prevent or select such deletions. This was demonstrated by the presence of mut-del2 in 42% of infected cells at 21 days post-infection, and mut-del1 in 54% at 13 days post-infection, in two of seven different cell cultures based on cells obtained from different donors.
Mut-del1 may be more important clinically than mut-del1, having been found in three of 68 specimens in an earlier study.
Conversely, the emergence of FCS deletions was suppressed in five other cultures, with less than 1% of deletions being found by day 13 post-infection. Thus, cultures of HAE-ALI from different donors led to the selection of different FCS deletions.
FCS deletions and viral entry
Mut-del2 does not lead to loss of the FCS, but prevents furin-mediated cleavage because of the upstream mutation. As a result, spike processing was dramatically impaired. Both deletions led to the inability to use either furin or TMPRSS2 to achieve fusion of the virus with the plasma membrane, thus inhibiting viral entry into the cell.
The range of cell tropism was also limited by FCS deletions. Cells that expressed TMPRSS2 did not show the presence of FCS deletions, supporting the role of the latter in abolishing the ability of the former to mediate virus-cell fusion.
However, FCS mutations among newly formed virions accumulated in two cell cultures. These were the same airway cell isolates that showed amplified levels of mutants, indicating their permissiveness to infection by FCS-deleted mutants. This could be because, lacking high TMPRSS2 expression, they allowed infection by other pathways that involved cathepsin-dependent endosomal entry of the virus.
Also, the findings of mut-del2-related reduction in viral entry and infection in Vero cells could indicate that FCS is a virulence factor. In this case, it should reduce the severity of lung disease in animals. Thus, further study is indicated to understand how often such deletions occur in human infection, how such mutants are transmitted between humans, and the extent of pathogenicity.
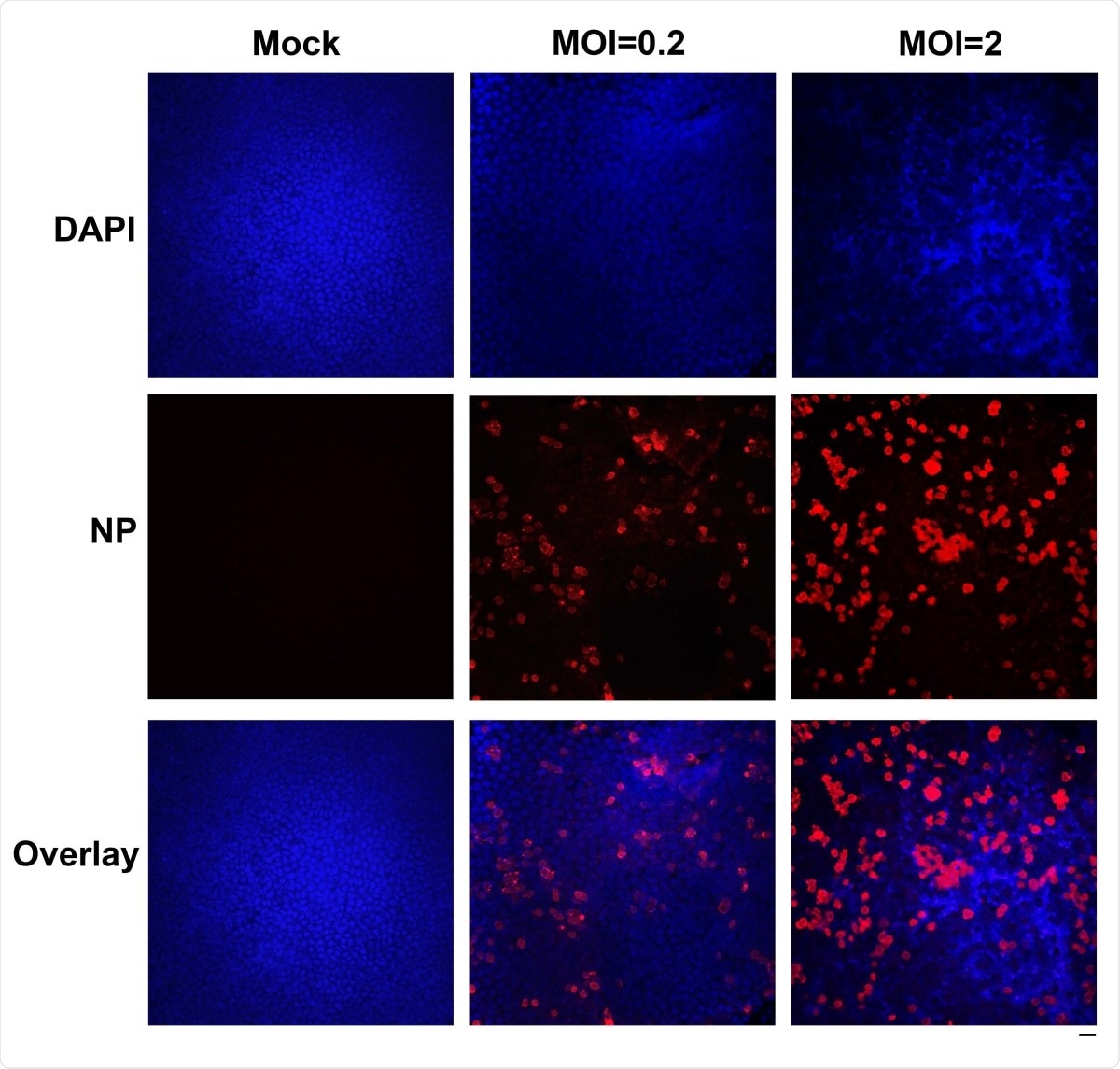
Immunofluorescence analysis of SARS-CoV-2 infection of HAE cells. HAE-ALIB2-20 cultures were infected with SARS-CoV-2 at an MOI of 0.2 or 2 pfu/cell, as indicated, mock-infected (Mock). At 4 days post-infection, a piece of the insert membrane was fixed in 4% paraformaldehyde in PBS at 4°C overnight. and subjected to direct immunofluorescence analysis. The membranes were stained with anti-SARS-CoV-2 N protein (NP). Images were taken on a Leica TCS SPE confocal microscope under 40×, which was controlled by Leica Application Suite X software. The nuclei were stained with DAPI (4’=,6- diamidino-2-phenylindole). Scale bar is 20 µM.
What are the implications?
These results demonstrate that the virus is subject to strong selective pressure within human airway cells, for adaptive mutations in the spike protein. Surveillance to identify spike mutations is essential to identify escape mutations, as well as those which affect or enhance viral infectivity or transmissibility, or virulence. The current study successfully used the polarized HAE-ALI model to explore spike mutants in a variety of scenarios.
The researchers found that the force of selection in viral particles released apically from infected polarized human airway cells led to stable retention of the FCS and overall spike stability in most cases, except for two cultures isolated from two different individuals. In these two cases, high FCS deletion rates persisted.
Our study presents evidence for the role of unique properties of human airway epithelia in the dynamics of the FCS region during infection of human airways, which is donor-dependent.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Zou, W. et al. (2021). The SARS-CoV-2 transcriptome and the dynamics of the 1 S gene furin cleavage site in 2 primary human airway epithelia. bioRxiv preprint. doi: https://doi.org/10.1101/2021.02.03.429670, https://www.biorxiv.org/content/10.1101/2021.02.03.429670v1
- Peer reviewed and published scientific report.
Zou, Wei, Min Xiong, Siyuan Hao, Elizabeth Yan Zhang, Nathalie Baumlin, Michael D. Kim, Matthias Salathe, Ziying Yan, and Jianming Qiu. 2021. “The SARS-CoV-2 Transcriptome and the Dynamics of the S Gene Furin Cleavage Site in Primary Human Airway Epithelia.” Edited by Xiang-Jin Meng. MBio, May. https://doi.org/10.1128/mbio.01006-21. https://journals.asm.org/doi/10.1128/mBio.01006-21.