Caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the coronavirus disease 2019 (COVID-19) pandemic has placed a significant burden on healthcare systems across the globe. To date, the virus has infected over 105.94 million people and led to over 2.3 million deaths worldwide.
Skyrocketing cases brought about by the second and successive waves have nearly overwhelmed healthcare systems in places like the U.S. and the U.K., underscoring the need for rapid diagnosis and treatment. Though vaccines are already being rolled out in many countries, global population immunity remains a long way off.
Researchers at the University of Nebraska Medical Center have developed a bacterial strategy for the expression and purification of the SARS-CoV-2 spike protein receptor-binding domain (RBD).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study, published on the pre-print medRxiv* server, showed that it is possible to rapidly and inexpensively produce functional and active antigens in bacteria to track and bind with the angiotensin-converting enzyme 2 (ACE2) receptor as well as antibodies in COVID-19 patient sera.
SARS-CoV-2 receptor-binding domain (RBD)
The SARS-CoV-2 virion contains four structural proteins – spike (S), envelope (E), membrane (M), and the nucleocapsid (N). The S protein works by protecting and covering the surface of the SARS-CoV-2 while allowing the virus to evade the host immune system and enter human cells. Attachment of the virus to the receptor relies on the receptor-binding domain (RBD), which is also responsible for inducing the production of neutralizing antibodies in the body.
Therefore, the RBD has been a highly-investigated target for vaccine and drug development. The interaction of the RBD and ACE2 receptor is an important event before viral infection. Past studies have shown that the RBD binds with the ACE2 receptor low nanomolar affinity, highlighting the key regions of the RBD that are responsible for ACE2 binding.
The RBD contains extensive β-sheets with an extended insertion called receptor-binding motif (RBM), which consists of the residues that make contact with ACE2. Apart from the β-sheets, the RBD also contains ten cysteines, which can stabilize the overall structure by forming five disulfide bonds.
The team noted that not all of these bonds are consecutively formed, while some need editing by reduction and reoxidation to generate the correct bonds, which can pose a complicated folding problem in the bacterial cytoplasm.
Production of recumbent proteins in bacteria
Attempts to obtain a recombinant protein using prokaryotic expression systems can be a fast and rewarding procedure, but it can be challenging. In most cases, the production of heterologous proteins in Escherichia coli strains has remained an experimental method.
Though the use of this bacteria is beneficial in terms of ease of production, cost-efficacy, and rapid growth, it can be a daunting task. It is vital to maintain the correct disulfide bonds for structural as well as functional stability, which can be problematic when expressing proteins of interest in the cytoplasm of E. coli.
The Ruddock lab developed the CyDisCo (cytoplasmic disulfide bond formation in E. coli) system that can produce active disulfide-bonded proteins in the cytoplasm of the bacteria without deleting reduction systems. It expresses the sulfhydryl oxidase Evr1p.
During protein folding, a new enzyme called protein disulfide isomerase (PDI) is added to edit disulfide bonds. The PDI can differentiate between properly folded proteins from misfolded ones, and correct disulfide bonds via cleave and formation cycles. Hence, properly and correctly folded proteins with disulfide bonds can be produced in the E. coli cytoplasm.
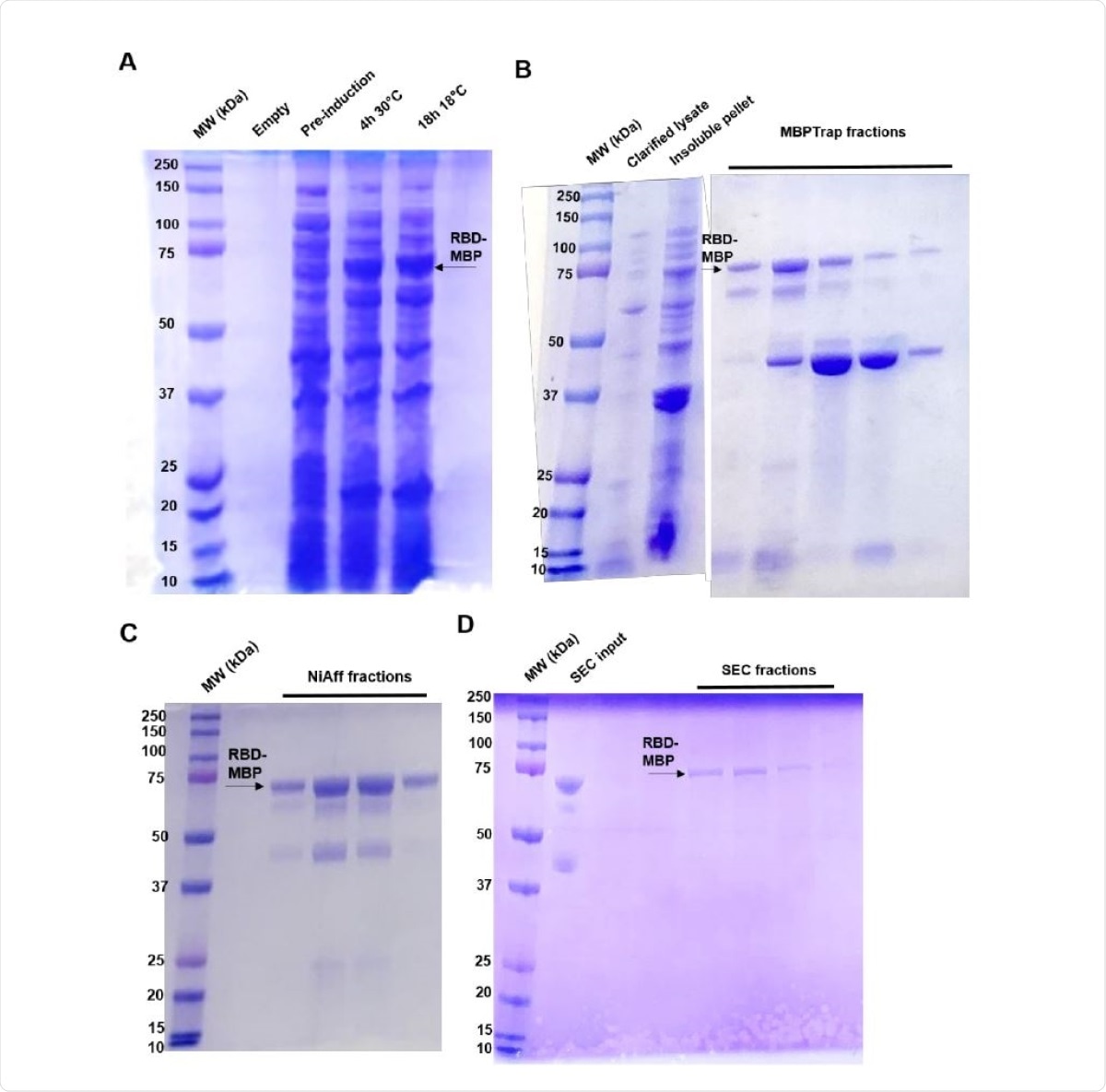
Protein purification SDS-PAGE gels. RBD-MBP can be observed running at 74 kDa, as indicated by the black arrows near the 75 kDa marker. Black arrows at approximately 42 kDa could indicate endogenously produced or cleaved MBP. A) Initial induction experiment comparing same-day (4 h) induction at 30° C and overnight (16 h) induction at 18° C. B) MBPTrap AKTA fractions. C) Nickel-affinity AKTA fractions. D) Size-exclusion AKTA fractions.
In the study, the researchers utilized the CyDisCo system to produce recombinant SARS-CoV-2 spike RBD that is properly folded with correct disulfide bonds. The team noted that the method is an easier and cost-effective way to generate antigens using a bacterial expression system, which can be used in diagnostic tests and assays.
To arrive at the study findings, the team developed a new and faster method to express and purify functionally active RBD antigen in E. coli. The team also found that the RBD fused to the maltose-binding protein (MBP) was recognized by human SARS-CoV-2-specific antibodies as confirmed by an immunoassay.
The coexpression of the sulfhydryl oxidase, Evr1p, and the disulfide bond isomerase, PDI in the CyDisCo system along with the plasmid of interest results in a well-folded and functionally active protein in a bacterial system, which is simple, rapid, and less expensive than a eukaryotic or mammalian system,” the researchers concluded in the study.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Prahlad, J., Struble, L., Lutz, W., et al. (2021). Bacterial expression and purification of functional recombinant SARS-CoV-2 spike receptor-binding domain. medRxiv. doi: https://doi.org/10.1101/2021.02.03.429601, https://www.biorxiv.org/content/10.1101/2021.02.03.429601v1
- Peer reviewed and published scientific report.
Prahlad, Janani, Lucas R. Struble, William E. Lutz, Savanna A. Wallin, Surender Khurana, Andy Schnaubelt, Mara J. Broadhurst, Kenneth W. Bayles, and Gloria E. O. Borgstahl. 2021. “CyDisCo Production of Functional Recombinant SARS‐CoV ‐2 Spike Receptor Binding Domain.” Protein Science 30 (9): 1983–90. https://doi.org/10.1002/pro.4152. https://onlinelibrary.wiley.com/doi/10.1002/pro.4152.