The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that is causing the current pandemic of coronavirus disease 2019 (COVID-19), has been the object of intense scrutiny and research over the last 14 months. Understanding its characterization is essential for the development of effective antivirals, vaccines or therapeutic antibodies.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
SARS-CoV-2 spike role
The virus attaches to and enters the host cell through its spike protein. This attachment involves the receptor-binding domain (RBD) on the spike protein, and the host cell receptor, the angiotensin-converting enzyme 2 (ACE2).
Both spike and receptor are glycosylated, the former much more heavily, with up to 22 N-glycans per protomer. The roles played by these sugars have been thought to include proper protein folding and conformational transformation, receptor attachment and epitope masking to avoid antibody neutralization.
Earlier studies have used pseudoviruses expressing the SARS-CoV-2 spike to understand the part this protein plays. Mutations at N-glycosylation sites have shown that the absence of these sugars at certain key residues in the RBD reduces infectivity markedly. Knock-out of the N-glycosylating gene MGAT1 also reduced the ability of a pseudovirus to accomplish invasion of the host cell. However, real SARS-CoV-2 cDNA clones have not yet been used to study this aspect.
N-glycosylation of spike
N-glycosylation is the primary post-translational modification occurring within eukaryotic cells. It begins in the endoplasmic reticulum (ER) membrane, the site of assembly of the molecule that acts as the universal precursor of all N-glycans, namely, lc3Man9GlcNAc2-dolichol-P. In a series of steps, the glycan is added to a growing peptide chain via the enzyme STT3, modified by α-glucosidases I and II within the ER, and thus exported to the Golgi apparatus.
After editing by α-mannosidases, and further structural changes to become a complex or hybrid glycan, it becomes a mature glycan, the exact structure being dependent on the type of cell and the species.
Several small inhibitors have been discovered that can act against enzymes along the entire pathway. Some are already in use to treat diabetes or type I Gauche disease, while others are being tested in clinical trials for broad-spectrum antiviral activity.
In the current study, the use of chemical or enzyme inhibitors or genetic knock-out, to disrupt N-glycosylation and the effect on infection in vitro was examined.
Chemical inhibition
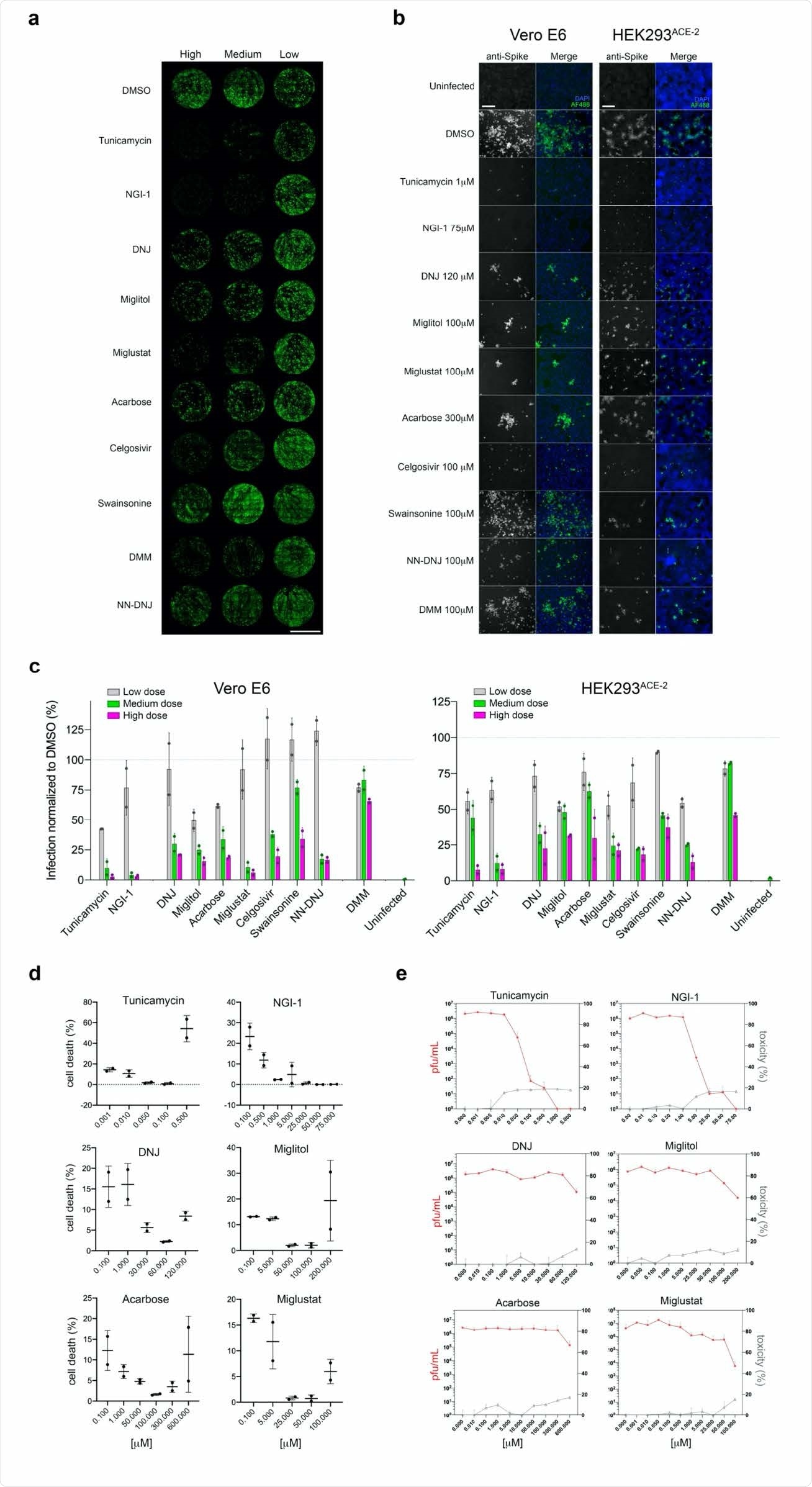
The researchers first used ten inhibitors acting along the N-glycosylation pathway. They found that these compounds mostly prevented infection with SARS-CoV-2 when added to the cell culture before, during or up to six hours after viral challenge. This was not due to direct cell toxicity, however, but due to the inability of the host cell to glycosylate the virus particles.
The effect varied between the inhibitors, with the most effective being those acting at the early steps of the pathway. These reduced infection by 92-98% in two types of cells, Vero and HEK (derived from African green monkey and human kidney epithelial cells, respectively). Inhibition of a-mannosidase I failed to prevent infection in either cell type to a significant effect.
Gene knockdown
The investigators used siRNAs to knock down genes encoding for key glycosylation enzymes before infecting the cells with SARS-CoV-2. Three knock-downs were performed, for the STT3 enzyme, the ER a-glucosidase I GANAB subunit, and the MGAT1 enzyme. The first knockdown reduced infection in both cell types by over 50%.
However, when different STT3 isoforms were separately knocked down, variable results were seen in Vero and HEK cells. With STT3-A and STT3-B, there was severe inhibition of infection in HEK and Vero cells, respectively. GANAB knockdown reduced infections in both cell types but to a lesser extent, at around 40% compared to ~50% with the STT3 inhibitors. Finally, MGAT1 knockdown showed a protective effect only in HEK cells, reducing infection by 40%.
Effects of non-glycosylation
Both the number of new virions, and the infected cell proportion, were reduced by almost 400 times following NGI-1 treatment relative to controls. Total viral RNA was only 80-fold less, which means that the alteration of or knock out of N-glycans on viral proteins led to the production of non-infective virions.
This was confirmed by using Peptide-N-Glycosidase F (PNGase-F), a compound that removes all N-glycans from glycoproteins. Cells treated with this compound remained uninfected, proving that SARS-CoV-2virions with non-glycosylated proteins were non-infective.
The researchers postulate that disruption of N-glycosylation prevents successful SARS-CoV-2 infection by interfering with viral protein folding or function; abolition of infection potential by newly formed virions; or mismatch of ACE2 receptors which are not properly folded.
Early inhibition most effective
Early inhibition of the pathway was found to be most effective, by preventing the formation of the universal precursor or its addition to the newly forming viral polypeptide. When STT3 knockdown included both isoforms, efficient protection occurred in both cell lines.
Later disruption of the pathway by ER a-glucoside I inhibition showed a lesser but still significant reduction in infectivity. There was also a significant difference in the inhibition achieved by different inhibitors. The iminosugars Miglustat, Celgosivir and NN-DNJ were most effective at reducing the viral load, the number of infected cells and the number of cells showing cytopathic effect.
The investigators attribute such variations in efficacy to differences in the N-glycosylation profiles between the host cell and the replicated virions.
Suppression of Infective virion production
Secondly, while control cultures showed infected cells forming clusters, suggesting viral spread between cells, the treated cultures showed infected cells as discrete entities. This may indicate that improper virion assembly, or failure of virion release, suppressing their ability to infect adjacent cells. In fact, the number of infected cells in treated samples always follows the initial dose of inoculated virus, while in controls, the cells show progressively increasing infection.
Cells with inhibited N-glycosylation could thus be incapable of producing glycosylated and, therefore infective virions, bringing the infection to an end. In fact, treating infected cells with a sub-inhibitory dose of NGI-1 reduced the number of infective virions compared to untreated cells, even though the amount of genetic material in both types of culture was similar.
Genetic ablation of host N-glycosylation genes reduces SARS-CoV-2 infectivity. (a) Representative whole-well scans of confluent Vero E6 monolayers transfected either with siNT (negative control) or siRNAs for the knockdown of the N-glycosylation genes siSTT3-A, siSTT3-B, siSTT3A+B, siGANAB and siMGAT1, and infected with SARS-CoV-2 (MOI=0.05, 24 hours). Cells immunostained with anti-spike antibodies (green), counterstained with DAPI (purple) and merged; scale bar 5 mm. (b) Representative images of either Vero E6 or HEK293ACE-2 242 siRNA cells infected with SARS-CoV-2 (MOI=0.05 for 24 hours for Vero; MOI=0.1 for 24 hours for HEK293). Cells immune-stained with anti-spike (gray channel) and merged (green) with DAPI counterstain (blue); scale bars 200 µm. (c) Quantification of the percentage of infected (spike-positive) cells in ‘a’normalized to siNT controls of either Vero E6 or HEK293ACE-2 cells infected with SARS-CoV-2 (MOI=0.05, 24 hours for Vero; MOI=0.1 for 24 hours for HEK293). Bars indicate the mean values; dots indicate individual values, error bars represent standard deviation; asterisks indicate significance (p<0.001); three technical replicates, four independent experiments performed. (d) Western blotting of protein lysates from Vero cells comparing siNT control cells with either siSTT3-A, siSTT3-B, siGANAB or siMGAT1 cells probing with specific antibodies against the knocked down protein, and with anti-GAPDH as loading control; apparent molecular weight markers annotated on the left.
What are the implications?
The study demonstrated that inhibiting the N-glycosylation pathway at important points led to a significant reduction in SARS-CoV-2 infection and replication in cell culture, using Vero and HEK cells. In most cases, the use of the inhibitor led to fewer infected cells, fewer cells showing the viral cytopathic effect, and reduced viral load in such cells. Gene knockdown studies confirmed these findings.
The most effective inhibitors prevented infection when added before, during or after infection. The lack or reduction in N-glycans prevents replicative infection with SARS-CoV-2.
Inhibitor-treated cells were producing a significant population of virions incapable of infecting further host cells, unlike untreated cells which produced infective virions only.”
This was confirmed by removing N-glycans from the surface of the intact virions, leading to a complete loss of infectivity. This indicates the essential role played by surface N-glycans in SARS-CoV-2 infection.
The evidence generated in this study opens the door to further research on developing ways to exploit the N-glycosylation inhibition and SARS-CoV-2 N glycans to treat COVID-19.”
Such therapies may be less vulnerable to escape mutations since they target a more conserved protein pathway.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.