As the coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to wreak havoc globally, the virus's exact source remains unclear.
Back in December 2019, a novel pneumonia-like illness was reported in Wuhan City, Hubei Province, China. By March 2020, the World Health Organization (WHO) had declared the global COVID-19 outbreak a pandemic.
Now, more than a year since it first emerged, the virus has reached 192 countries and territories. To date, more than 109.14 million cases have been confirmed, and over 2.40 million have lost their lives.
As the exact zoonotic origin and evolution of SARS-CoV-2 remain unresolved, scientists at the Francis Crick Institute in London, UK, explored the structure and binding properties of a Pangolin coronavirus spike glycoprotein to see if it could shed light on the evolution of SARS-CoV-2.
SARS-CoV-2 source
Since the virus was first reported, many scientists have been exploring its zoonotic and phylogenic origins. Like previous outbreaks of betacoronaviruses (in the same subgenus as SARS-CoV-2) – such as the severe acute respiratory syndrome (SARS) in 2002 in China and the Middle East Respiratory Syndrome (MERS) in 2012 in Saudi Arabia – horseshoe bats were considered the likely source. From there, the virus is believed to have jumped from an as yet unidentified intermediate host.
Coronaviruses of bats and pangolins have been implicated in the origin and evolution of the COVID-19 pandemic. As aforementioned, SARS-CoV-2 belongs to the subgenus of sarbecoviruses, which thrive in both horseshoe bats and pangolins.
Through a previous study, the current research team demonstrated that the bat coronavirus RaTG13, the closest known relative of SARS-CoV-2, is unlikely to infect human cells since its spike protein has a low affinity for the human angiotensin-converting enzyme 2 (ACE2) receptor.
As a result, it has been theorized that SARS-CoV-2 could have jumped to humans through an intermediate host. Several studies reported the existence of sarbecoviruses highly similar to SARS-CoV-2 in diseased Malayan pangolins. As such, pangolins were considered to have played a role in the emergence of the pandemic.
ACE2-binding properties
In the study, published in the journal Nature Communications, the team analyzed ACE2-binding properties and S protein structure from a Pangolin coronavirus (CoV) closely related to SARS-CoV-2.
To arrive at the study findings, the researchers expressed and purified two various Pangolin-CoV spike ectodomains, which can characterize the pangolin virus spike and compare it with that of SARS-CoV-2. The team based these on sequences of viruses isolated from pangolins seized in China's Guangdong province in 2019.
Further, the team generated ectodomains of ACE2 proteins from humans, bats, and pangolins to perform comparative assays.
The team has found that pangolin proteins demonstrated strong binding to the human ACE2, about ten-fold weaker binding to pangolin ACE2, and fragile binding to bat ACE2. A similar pattern of binding was noted for SARS-CoV-2, which had a strong binding to human ACE2.
The team added that the Pangolin-CoV S structure is similar to the closed-form of the SARS-CoV-2 S and the RatG13 S.
All of the observations suggest many sequence-based differences that make the Pangolin coronavirus spike adopt an all-closed conformation. The Pangolin-CoV S's closed conformation is remarkably similar, outside of the receptor-binding domain (RBD), to the RaTG13 S, suggesting that the latter's structure was potentially not materially impacted by the crosslinking.
The scientists believe that the closed conformation role was for shielding the fusion apparatus, while the open confirmation status is required to facilitate receptor binding.
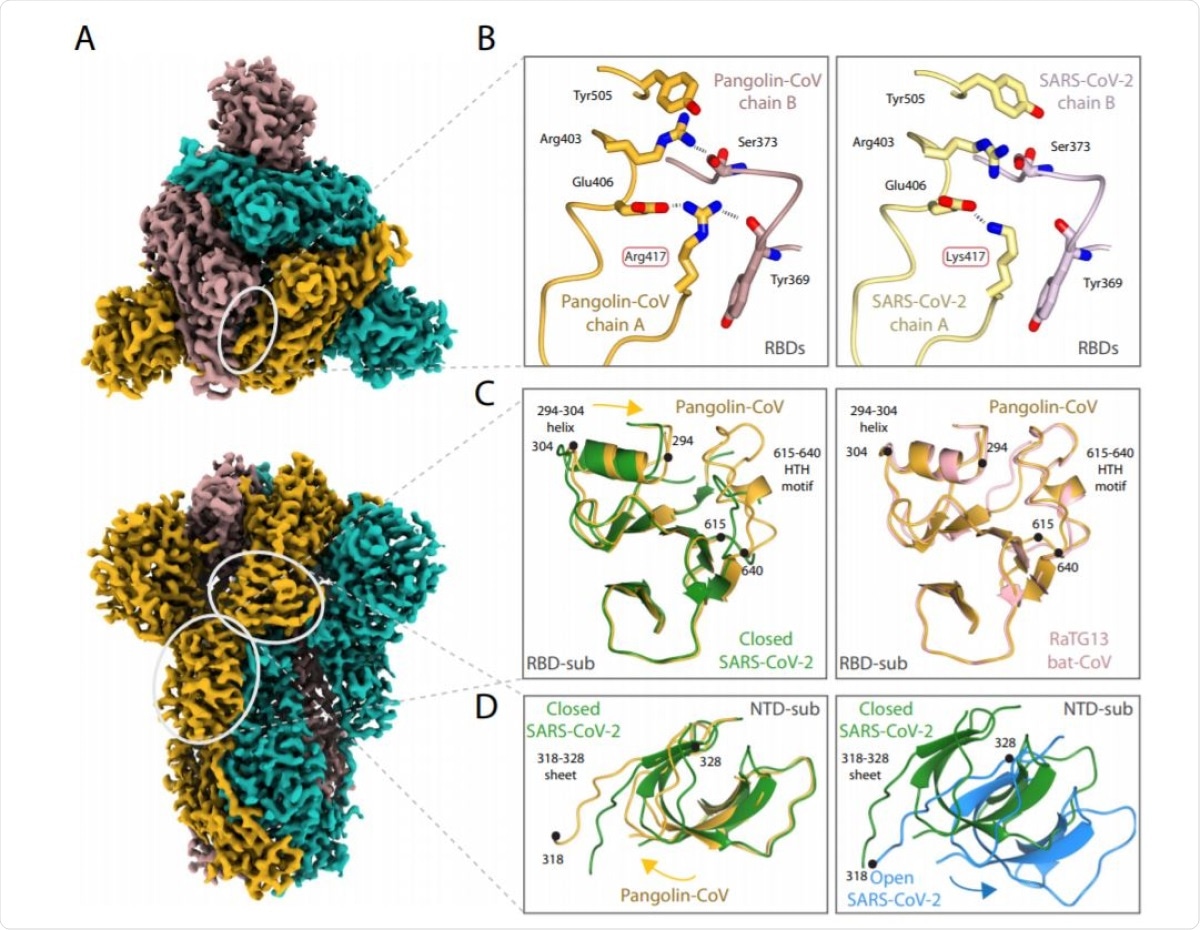
Structure of Pangolin-CoV spike protein. A EM density representation from the 2.9 Å map of Pangolin-CoV S viewed from down the three-fold axis (top panel) and in the orthogonal view (lower panel). The subunits are coloured in sea blue, golden and rosy brown. The white ovals identify the areas shown in molecular representation on the right. B Comparison of the RBD/RBD interface from the pangolin-CoV (left) and SARS-CoV-2 S (PDB: 6ZGE, right) highlighting the Arg417Lys substitution. C Comparison of the RBD-associated subdomains of the pangolin-CoV (golden) and closed form of SARSCoV-2 (green) in the left hand panel, showing the different positioning of the 294–304 helix and the presence of the 615–640 helix-turn-helix in the pangolin structure and, in the right hand panel, the overlap of the same Pangolin-CoV S structure (golden) with the corresponding region from the RaTG13 (PDB: 6ZGF) (pink). D Comparison of (left) the NTD-associated subdomain of Pangolin-CoV (golden) with that of the closed form of SARS-CoV-2 (green) showing the different domain orientations between them; (right) the closed (green) and open (blue) conformations of the NTD-associated subdomain of SARS-CoV-2 showing that the shift in orientation of the NTD-associated subdomain on spike opening is in the opposite direction to the shift seen between the Pangolin-CoV and closed SARS-CoV-2 conformations shown in the left panel.
The study also showed that the presence of ACE2 receptors enhances the opening of the RBDs of the SARS-CoV-2 spike and its priming for succeeding membrane fusion. An open conformation of the SARS-CoV-2's spike may help create an early kinetic event in the binding process.
Further, the SARS-CoV-2 S protein non-RBD component is comparable to that of the bat virus RaTG13 protein, with a 96 percent identity with S1. Meanwhile, their sequence identity is just 76 percent in the RBD.
Contrarily, the sequence and SARS-CoV-2's RBD is similar to that of Pangolin-CoV, mainly at the ACE2-binding site. The close similarity of the RBD between Pangolin-CoV and SARS-CoV-2 links to the near-identical binding properties of their two S proteins.
The researchers suggest that the effective zoonotic range for this class of coronaviruses beyond bats may include other species like pangolins. These animals have ACE2 receptors akin to the human ACE2. There might be unidentified pathogens like viruses that harbor RBDs of similar sequence and binding properties to SARS-CoV-2 and Pangolin-CoVs.
Understanding these similarities and differences can also help scientists explore other potential host animals, which can, in turn, guide preventative measures for similar zoonotic spill-overs in the future.