Even as vaccination efforts are underway to achieve population immunity against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic, disturbing evidence is coming to light that the emergence of newer strains may hinder this desirable outcome.
A new study by researchers in France reports impaired neutralization of two new SARS-CoV-2 variants by antibodies induced by vaccination or natural infection. The team has released their findings on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Important variants
The first widely recognized variant was the D614G mutation in the viral spike protein. This was found to confer a transmission advantage by stabilizing the conformation of the spike towards a state favorable for binding the host angiotensin-converting enzyme 2 (ACE2) receptor.
Towards the latter part of 2020, the so-called British variant was identified, followed shortly by the South African and Brazilian variants (the B.1.1.7, B.1.351 variant or 501Y.V2, and the P.1 and P.2 lineages, respectively). These show combinations of mutations in the spike region, including the receptor-binding domain (RBD) and N-terminal domain (NTD), as well as other genomic regions.
It is noteworthy that all of them share some escape mutations already known to have emerged under the selection pressure of antibody exposure, both from immune serum or therapeutic antibody use in prolonged survival within immunocompromised individuals, and from the antibodies elicited by natural infection.
However, all do not show the same level of immune evasion, with the UK variant being more susceptible to neutralization than the other two. The well-known N301Y mutation by itself does not appear to be responsible for resistance to neutralization by vaccine-induced antibodies.
The mutations in both the B.1.351 and P.1 strains, including E484K and K417N/T, are thought to enhance viral fitness and may confer antibody resistance in part, at least.
Earlier studies showed that the UK variant spike-bearing pseudovirus was neutralized by the antibodies elicited by the Pfizer vaccine, but with somewhat lower titers relative to the reference Wuhan strain. Likewise, the Moderna vaccine antibodies suffered a 5-10-fold loss of efficacy against the South African spike compared to the D614G pseudovirus.
When assayed using the authentic B.1.351 virus, however, neutralization by plasma from six convalescent donors was markedly weaker, with the 50% inhibitory concentrations (IC50) rising by 6-200 times relative to the original strain. This makes it a high priority that standardized assays using authentic virus be used in addition to pseudoviruses, to measure the sensitivity of the virus to antibodies.
Study details
The current study looks at the neutralization of three strains of the virus; namely, the D614G virus and the B.1.1.7 and B.1.351 variants. The researchers used two anti-RBD antibodies, mAb102 and mAb48, that bind to the RBDs in the ‘up’ conformation and thereby prevent RBD-ACE2 binding.
While mAb102 was able to neutralize all three strains at low titers, mAb48 neutralized only the D614G strain, showing that antibody sensitivity varies with strain.
The three variants were incubated with serum from a range of convalescent COVID-19 subjects, who had mild, moderate and severe-to-critical disease. Samples were taken at month 3 and month 6 from the onset of symptoms (M3 and M6, respectively).
B.1.351 strain partly resistant
At both time points, D614G and B.1.1.7 strains showed equal sensitivity to the sera, but with large inter-individual variations over two orders of magnitude. Neutralizing activity was more potent in sera from those with critical disease relative to all others, but was maintained in all cases at six months.
Neutralization was five-fold and ten-fold weaker when the sera was tested against the B1.351 variant at M3 and M6, respectively, relative to the other two strains.
These results were validated by another experiment involving serum from people with mild disease, collected much later, at 233 days from the onset of symptoms. This showed lower neutralization activity, with IC50 values being equivalent for both D614G and B.1.1.7 strains. However, with B.1.351, the neutralizing potency was four-fold less compared to D614G.
While most of the sera neutralized all three strains at M1, there was a noticeable drop by M6, affecting the B.1.351 strain the most. The second study showed a higher proportion of neutralization at 93%, against either the D614G or B.1.1.7 strains, at M9. With the B.1.351 strain, only 63% of sera achieved neutralization.
Binding affinity lost
When tested against the spike antigen for binding, mAb102 bound the spike protein of all three variants equally. The mAb48 bound with high affinity to the D614G spike, somewhat less efficiently to B.1.1.7, and not at all to B.1.351, indicating that changes in antibody binding mediate mutational antibody escape.
The N501Y mutation is found to increase the RBD-ACE2 binding affinity. In the current study, soluble ACE2 was bound to a far greater extent to B.1.1.7, compared to D614G, and also to B.1.351, though the increase was not so dramatic in this case.
When tested against the same convalescent sera, both B.1.351 and B.1.1.7 showed higher affinity for the ACE2 receptor, more with the latter, but were not neutralized by some monoclonal antibodies, and even less, or not at all, with polyclonal sera.
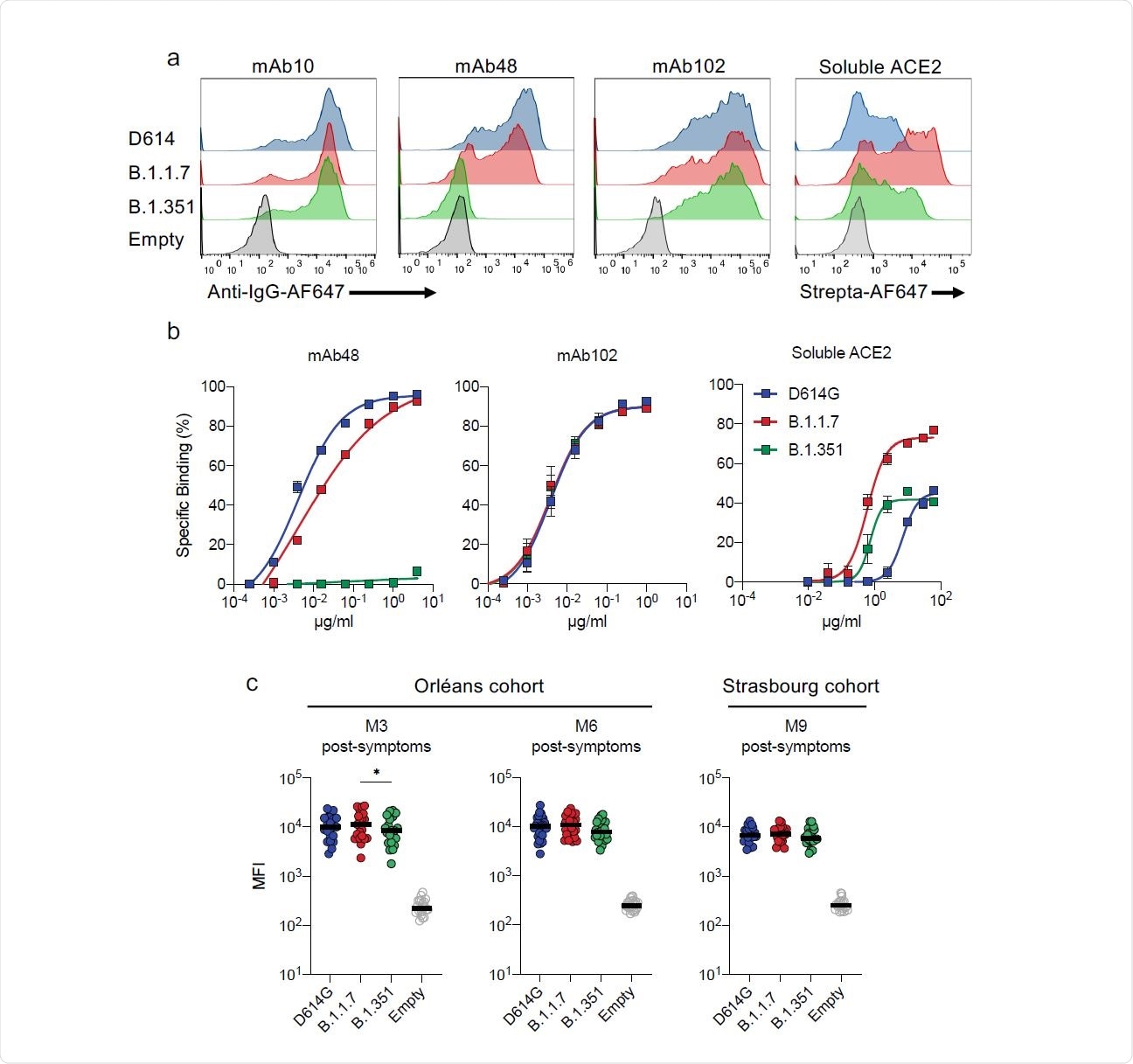
Antibody binding to cells expressing D614G, B.1.1.7 and B.1.351 Spikes. a. Binding of monoclonal antibodies or soluble ACE2. 293T cells were transiently transfected with plasmids expressing the D614G, B.1.1.7 and B.1.351 Spikes. After 24h, cells were stained with anti SARS-CoV-2 antibodies mAb10 (a pan-coronavirus antibody), mAb48, mAb102 or soluble ACE2 (ACE2- biotin at 10 µg/ml revealed with fluorescent Streptavidin) and analyzed by flow-cytometry. One representative example of binding is shown. b. Titration binding of curves of mAb48, mAb102 and ACE2 to the three Spikes. Data are mean of three independent experiments. c,d. Binding of the panel of 83 sera from 58 convalescent individuals. Sera were tested at a 1/300 dilution. Data are mean of two independent experiments.
Vaccine-induced antibodies show low, delayed response against B.1.351
When the sera from 19 Pfizer vaccine recipients was tested against the three strains, most of them being SARS-CoV-2-naïve, they found that at week 2, only D614G was neutralized. At week 3, neutralizing activity began to be detected against B.1.1.7 as well but with lower potency.
At week 4, both these strains showed equal neutralization. The B.1.351 strain escaped neutralization until week 3, becoming discernible only at week 4, but still was less efficiently neutralized than the others.
With low antibody titers at week 2 post-vaccination, D614G neutralization occurred with only 5/15 sera, and B.1.1.7 neutralization with only 2 sera, and none against the third strain. At week 3, both the former strains were neutralized by 63% and 38% of sera, respectively, but with a three-fold decline in neutralization titers against the second strain. B.1.351 continued to be resistant.
Neutralizing titers increased a week after the second dose, with 80% of sera achieving neutralization against D614G and B.1.1.7. The neutralizing titers were comparable for these two, but only 60% neutralization was achieved, at seven-fold lower titers, for B.1.351.
This indicates that the Pfizer vaccine neutralizes D614G and B.1.1.7 efficiently, with some efficacy as little as two weeks after the first dose, but with a delay against the third strain, accompanied by lower neutralizing titers.
“Our results suggest that the low neutralizing titers (ED50 of 50-100) in the sera correspond to a protection against severe disease.”
Nasal swabs inactive against B.1.351
Nasal swabs in immunized patients were tested for mucosal immunity leading to neutralization of SARS-CoV-2. While three vaccine recipients had signs of prior infection, they showed high neutralizing titers at week 2, and neutralizing activity in the nasal swabs against D614G and B.1.117 strains but not B.1.351.
Overall, of the 17 swabs from vaccine recipients, neutralizing antibodies were found in 6, 7, and 8 swabs at weeks 2, 3 and 4, respectively. Mucosal immunity is thus weak soon after vaccination.
What are the implications?
The study suggests that the B.1.351 variant is resistant to natural or vaccine-induced antibodies in a large proportion of subjects, especially at low titers. This is mediated by the E484K mutation, found in both the B.1.351 and P.1 variants, and recently in the UK variant too, posing a major challenge for population immunity. The use of authentic virus allowed viral fitness and the impact of other mutations on neutralization to be evaluated.
The study also shows a convenient and rapidly adaptable workflow that can be used for the rapid analysis of any emerging variant. Further work will help reveal the roles played by local and systemic antibodies in the protective efficacy of vaccines. Poor antibody responses can thus help predict the loss of cross-reactivity of a newly emerging strain to pre-existing humoral immunity.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Planas, D. et al. (2021). Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. bioRxiv preprint. doi: https://doi.org/10.1101/2021.02.12.430472, https://www.biorxiv.org/content/10.1101/2021.02.12.430472v1
- Peer reviewed and published scientific report.
Planas, Delphine, Timothée Bruel, Ludivine Grzelak, Florence Guivel-Benhassine, Isabelle Staropoli, Françoise Porrot, Cyril Planchais, et al. 2021. “Sensitivity of Infectious SARS-CoV-2 B.1.1.7 and B.1.351 Variants to Neutralizing Antibodies.” Nature Medicine, March. https://doi.org/10.1038/s41591-021-01318-5. https://www.nature.com/articles/s41591-021-01318-5.