A team of researchers at Georgetown University Medical Center, USA, investigated how severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) affects the immune system’s response during and after infection and contributes to the severity of coronavirus disease 2019 (COVID-19).
Not only did the team observe that SARS-CoV-2 altered the immune response through several signaling molecules and genes, but they also found genetic changes linked to an increased risk of cancer.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study “Immune characterization and profiles of SARS-CoV-2 infected patients” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
Methods
The team profiled the immunological response of patients with severe COVID-19 infection. They analyzed different RNA sequence profiles collected from nasopharyngeal swabs that collected viral load from the upper respiratory tract of 430 patients with COVID-19 infection. They also collected data on 54 patients who did not SAR-CoV-2 for a baseline of the immune response. Gene-specific analyses were performed based on the information from four public databases from the NCBI GEO database.
Pathway analyses of differentially expressed genes were also executed to characterize modulating proteins needed for COVID-19 infection. The researchers suggest future medications targeting these pathways can help make future therapeutic strategies for alleviating severe infection.
Immunological profiles of patients with SARS-CoV-2
Genetic results showed a downregulation of genes in patients with SARS-CoV-2, with the most downregulated genes being DPM3, ROMO1, MT3, MRPL53, SCGB3A1. Although, the above protein-coding genes are not directly linked to the immune response. The authors suggest the infection’s inhibitory effect on the upper respiratory tract may not be specific to the immune system but, instead, may cause widespread damage to other processes that then overlap with immune function.
Genes that were upregulated by SARS-CoV-2 were directly associated with the immune system, including interferon-induced proteins IFI44L, IFIT1, OAS3, and RSAD2 involved in antiviral response.
Based on 17 patients with COVID-19 infection and 17 who were uninfected, SARS-CoV-2 upregulated IGHG1, IGHV4-59, IGHV1-24, IGHV1-46, IGKC, and downregulated CACNA2D3, LINC00877, DHRS9, and CASC8.
SARS-CoV-2 alter gene expression in signaling pathways
The researchers found the eIF2 signaling pathway had the most downregulated genes due to COVID-19 infection. “The stress response induces translational shutdown via the phosphorylation eIF2α. Therefore, sustained phosphorylation of eIF2α inhibits host or viral protein synthesis. Our IPA core analysis shows eIF2α downregulated in the eIF2 signaling pathway, as well as other mediators of the eIF2 complex, including eIF2γ.”
The researchers note that it remains unknown which upstream regulator of the stress response influences COVID-19’s downregulation of the eIF2 signaling pathway. However, they suggest PERK and PKR could be potential targets for future treatment to prevent the coronavirus from taking over the host cell’s replication machinery and also regulate interferon response.
Results also showed the downregulation of genes MRPL53 and ROMO1 in pathways involved in mitochondrial dysfunction and oxidative phosphorylation.
Mitochondrial dysfunction may be an indirect cause of the lymphopenia that is often observed in severe COVID-19 patients,” wrote the authors. “While the analysis of downregulated genes in this RNA-profile does not directly relate to the immune response, there is a relationship between these genes and the host-viral interaction, which requires further investigation.”
Upregulation of genes was observed in the pathway involved in an innate immune response, including interferon-alpha as an upstream regulator. The authors suggest the innate immune response cannot ward off SARS-CoV-2 because of a dysregulated interferon response.
Genetic changes influence disease severity
In patients with severe infection, pathways such as the Hepatic Fibrosis Signaling, Osteoarthritis, and Neuroinflammation pathways were activated in a COVID-19 infection while interferon signaling was inhibited. “This suggests that these pathways contribute to the severity of COVID-19 disease.”
In addition, patients with severe COVID-19 infection had upregulated MX1, IFIT1, and HLA-DQA2 with IFIT1 and MX1 “more enriched” compared to patients with mild infection.
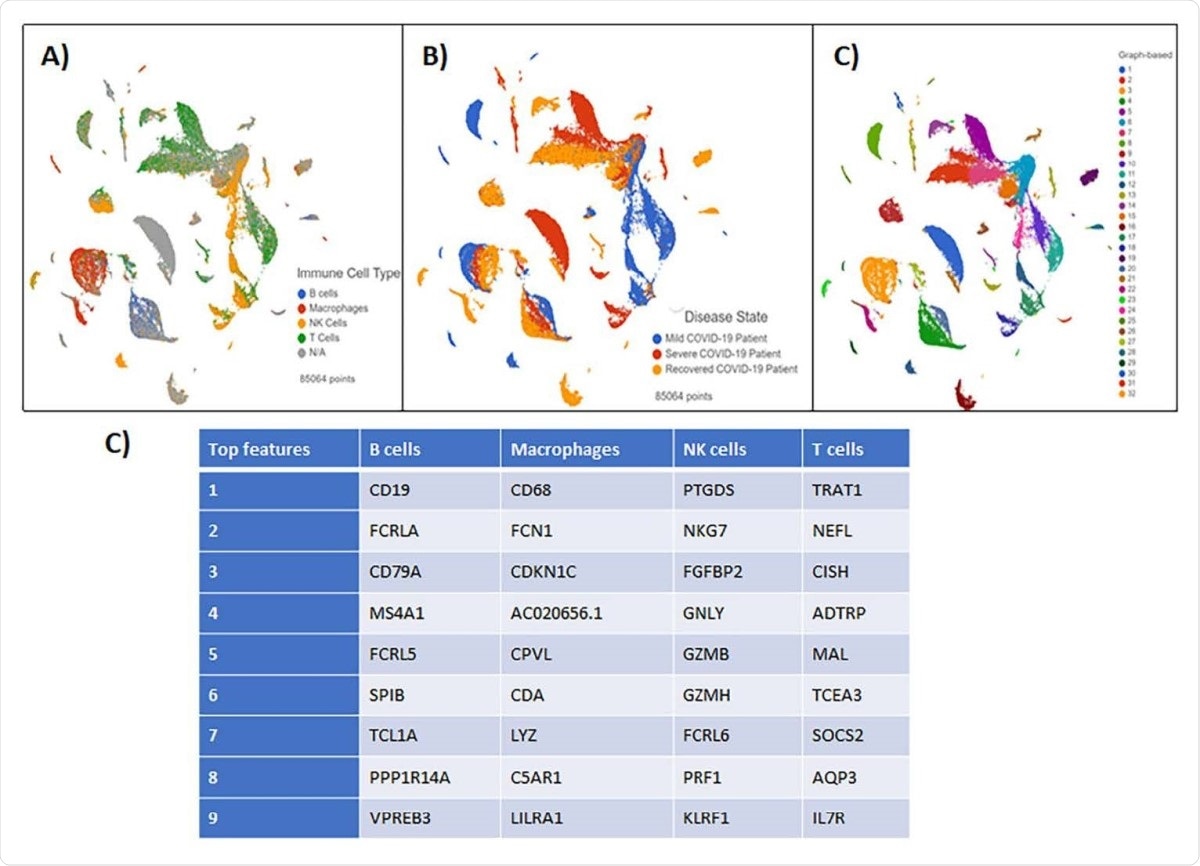
Blood Buffy Coat Clustering Analysis results visualized by Global UMAP (A) Patient samples were grouped by disease severity. (B) Cells were classified by four immune cell subsets: B-cells, Macrophages, NK cells, and T cells. N/A defines all unclassified cells. (C) Graph-based analysis was performed in Partek Flow. 25 clusters were reported (D) The top features generated by clustering analysis in each immune cell subset.
Patients who recovered from infection displayed regained expression of the HLA-DQA2 host factor along with 10 other genes. However, patients with severe infection showed the same level of upregulated and downregulated genes. “Our analyses for severe COVID-19 patients suggest that upregulated interferon-induced proteins, chemokines, and cytokines contribute to the severity of COVID19 compared to mild patients.”
SARS-CoV-2 dysregulates cells which may increase cancer risk
A major finding of the study was the relationship between the coronavirus pathogenesis pathway with the transcription factors CSF2 and E2F7. Gene downregulation of transcription factors Pax6, RB1, and RELB, may cause alterations in the cell cycle and cause oncogenes. The authors suggest an indirect effect of genetic changes caused by SARS-CoV-2 may increase the risk for cancer.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources