To better understand the transient and chronic autoimmune symptoms caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, researchers from Memorial Sloan Kettering Cancer Center, Guizhou Medical University, and biotech company Curandis embarked on an endeavor to establish a comprehensive autoantigenome for COVID19.
In a previous study, the team identified a repertoire of autoantigens (autoAgs) from human fetal lung fibroblast HFL1 cells strongly tied to neurological and diverse autoimmune symptoms COVID19.
In the current study, they aimed to identify more autoAgs from A549 cells, which are human lung epithelium-like cells, derived from adenocarcinoma cells used to model SARS-CoV-2 infection.
The unique binding affinity between the autoAgs and dermatan sulfate (DS), a glycosaminoglycan, was exploited to identify the former. Their binding leads to robust dual B cell signaling in B1 cells reactive to host antigens, thus eliciting autoantibodies.
Any self-molecule that binds to DS thus has a significant potential for autoantigenicity, pointing to how a diverse array of self-molecules trigger the same or highly similar immune cascades, leading to autoimmune antibody generation.
Autoantibodies in SARS-CoV-2 infected cells
Autoimmune phenomena in COVID-19 include multisystem inflammatory syndrome in children, immune blood cell disorders, antiphospholipid syndrome, immune-mediated neurological syndromes, and Guillain-Barré syndrome. Both the classical autoantibodies antinuclear antibody (ANA) and extractable nuclear antigen (ENA) are found in some COVID-19 patients, as well as others including anti-neutrophil cytoplasmic antibody, lupus anticoagulant, antiphospholipid, and anti-interferon (IFN) antibodies.
The source of these autoantibodies is virally mediated modification of the co-opted host cellular components that cause self-molecules to change into autoAgs. This infection's molecular mechanisms include direct interaction with viral proteins, disruption by expressed viral proteins, and post-translational protein modification by virally induced ubiquitination and phosphorylation.
The researchers found 348 proteins from A549 cell extracts, 214 with strong and 134 with moderate DS-binding affinity. They showed around three-fold higher interactions with each other, compared to the expected 2.500 protein-protein interactions (PPIs). This is a significant indicator that they belong to common biological pathways.
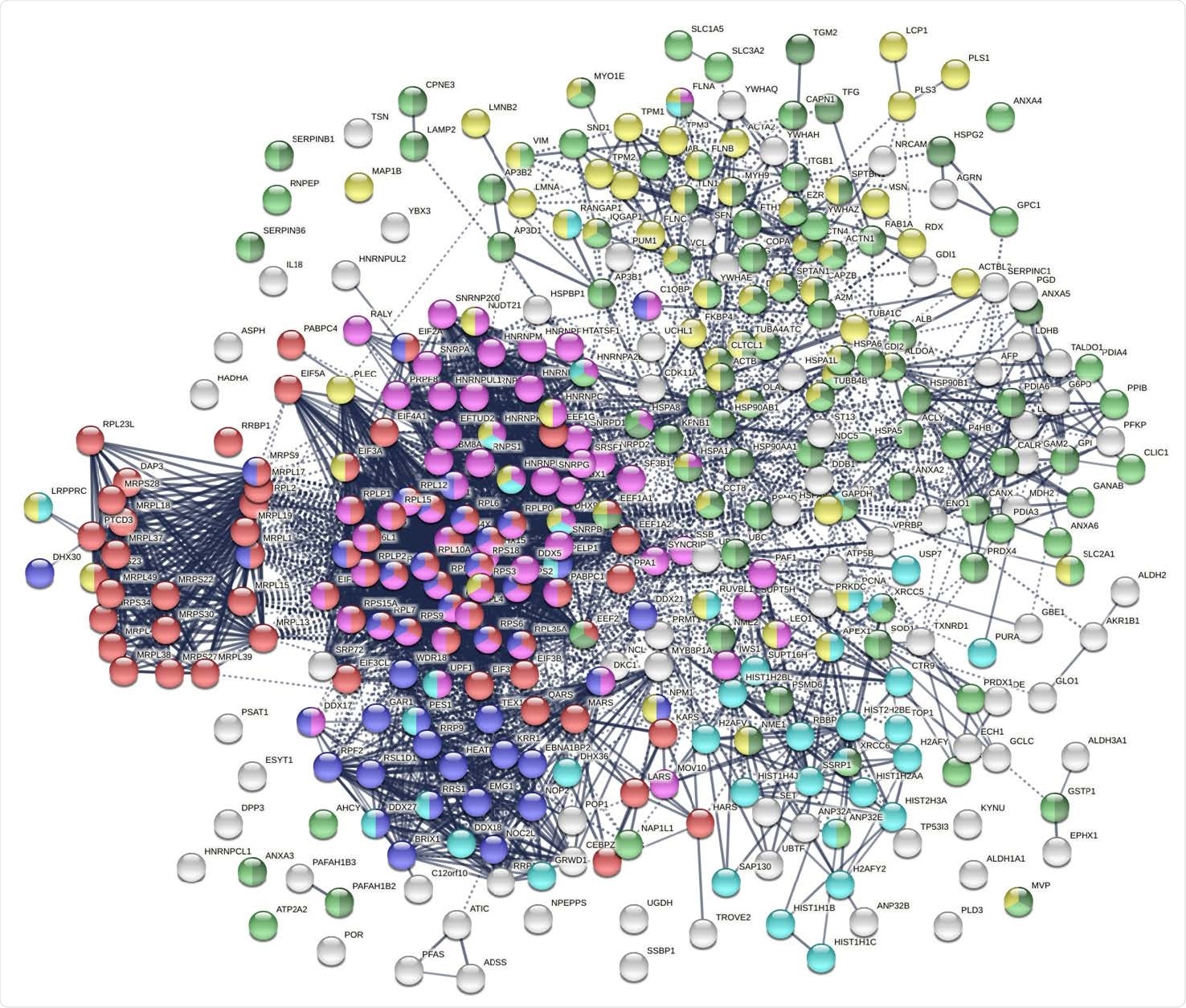
The autoantigenome from A549 cells identified by DS affinity. Marked proteins are associated with translation (69 proteins, red), mRNA metabolic processing (69 proteins, pink), ribosome biogenesis (43 proteins, blue), vesicle (87 proteins, green) and vesicle-mediated transport (72 proteins, dark green), chromosomes (40 proteins, aqua), and cytoskeleton (65 proteins, gold).
SARS-CoV-2-altered proteins
The researchers then compared the autoAg-encoding genome with the COVID-19 transcriptome to find that 84% of the 348 autoAgs showed altered mRNA or protein expression. The discrepancies (either up-regulation or down-regulation) are likely to be due to the use of different techniques and cell lines in various studies.
About 66% of the altered proteins were autoAgs already reported in earlier studies. The same increased network of interactions was found with the COVID-19 altered proteins, indicating involvement with ribonucleoprotein complex formation.
Other related pathways include protein translation, mRNA metabolic processes, ribonucleoprotein complex formation, and vesicle trafficking.
AutoAgs through SARS-CoV-2 PPIs
The study shows that 25/38 DS-affinity proteins that take part in PPIs with SARS-CoV-2 proteins are known autoAgs, especially ORF3 (open reading frame 3) that interacts with nine DS-binding proteins and six autoAgs. The viral nucleoprotein (N) antigen and non-structural protein (NSP)-4 also show such PPIs.
AutoAgs formed by perturbed viral protein expression
While almost 170 autoAgs were produced by perturbation of the expression of viral proteins, the most deeply involved was ORF3. Over 70 DS-binding autoAgs were produced by the perturbation that caused this single protein. This suggests the crucial role played by ORF3 in SARS-CoV-2 infection.
Others include the envelope (E) protein, ORF9b, and the membrane (M) protein. Classical nuclear autoAgs are also observed, as are platelet-activating factor (PAF) type I. The regulation of PAF by ORF3 could underlie, in part, the thromboembolic complications so often seen in COVID-19 patients.
Mitochondrial perturbation – viral ORF9b
Over a score of mitochondrial ribosomal proteins with strong DS affinity were found in SARS-CoV-2 infection, most of them being downregulated. Eleven of the 16 proteins downregulated by ORF9b were from this protein category, indicating that mitochondrial protein synthesis could be affected. The SARS-CoV-2 infection could, therefore, primarily affect the mitochondria via ORF9b expression.
Interestingly, this protein also suppresses type I IFN responses and perturb mitochondrial structure and function. Mitochondria within monocytes of COVID-19 pneumonia patients thus lose their ability to maintain energy homeostasis. These changes may help evade the host immune response and produce muscle weakness and fatigue due to altered energy metabolism.
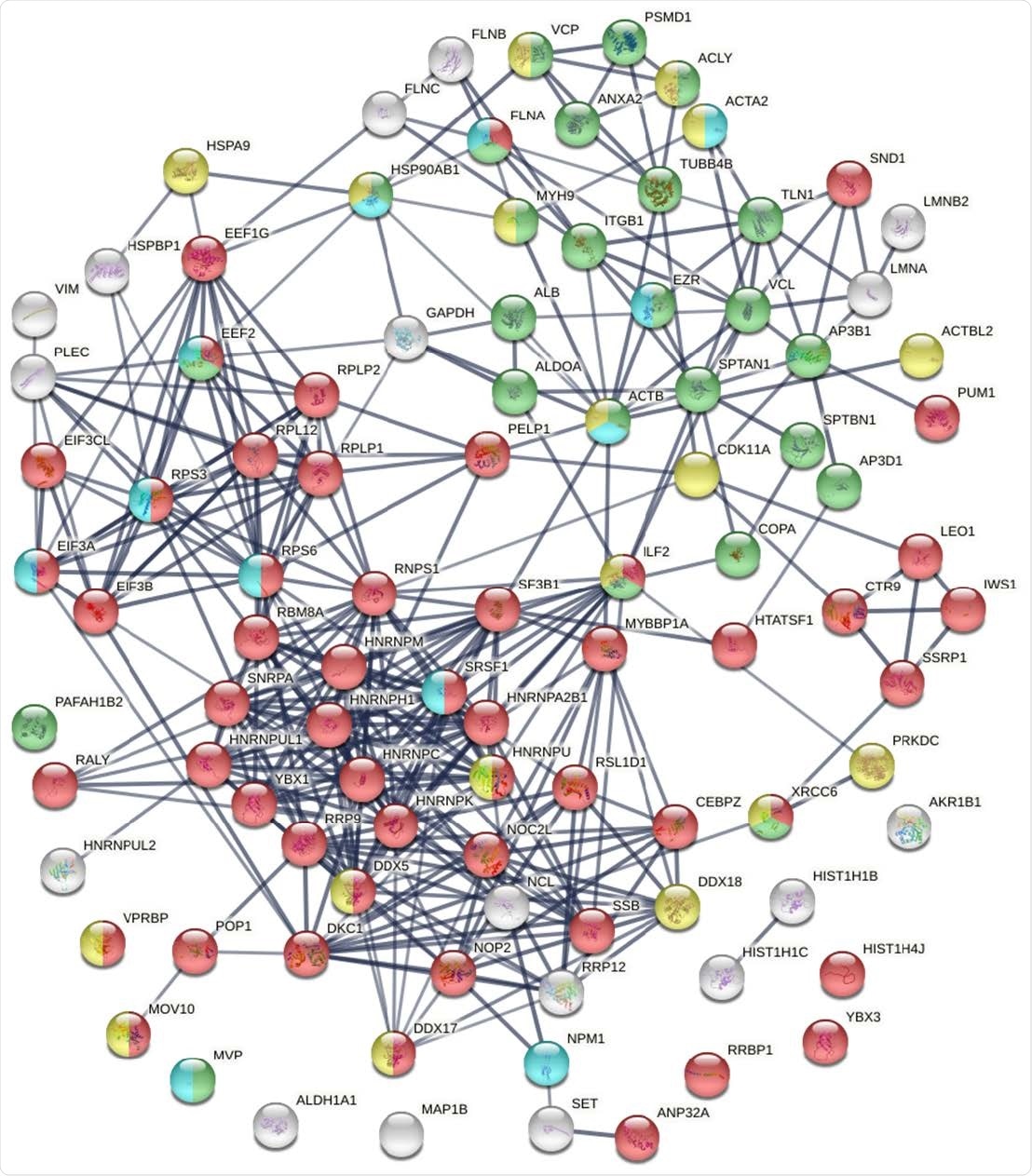
Known and putative autoAgs derived from phosphorylation alteration in SARS-CoV-2 infected cells. Marked proteins are associated with gene expression (52 proteins, red), vesiclemediated transport (25 proteins, green), ATP binding (18 proteins, gold), and kinase binding (12 proteins, aqua).
Ubiquitination alterations and phosphorylation
Ubiquitination alterations in SARS-CoV-2 infection lead to increased protein breakdown and are identified with ten altered DS-binding proteins in COVID patients. Others, like papain-like proteases (PLP), also reverse or prevent ubiquitination and remove IFN-stimulated gene product 15 (ISG15).
These changes affect the host immune response, and a large number of such proteins in this infection could indicate that this is a significant source of autoAgs in COVID-19.
Phosphorylation of proteins during apoptosis often leads to their being targeted by autoantibodies. Dephosphorylation of some self-molecules also leads to autoAg formation.
Conclusion
The study reports the identification of 191 confirmed autoAgs and another 150 possible autoAgs in COVID-19 patients. First detected in the human lung epithelial cell line A549, by DS affinity-based enrichment, they were compared against COVID proteomics and transcriptomics.
This shows that autoAgs are formed from many cell processes and components hijacked by SARS-CoV-2 during the infection. Mitochondrial perturbation and post-translational modification, ubiquitination, and phosphorylation are involved in autoAg formation.
“Overall, our study demonstrates that SARS-CoV-2 causes extensive alterations of host cellular proteins and produces a large number of potential autoAgs, indicating that there may be an intimate relationship between COVID infection and autoimmunity.”