A study recently conducted at the Jenner Institute, University of Oxford, UK, has investigated the significance of a transmembrane glycoprotein basigin, or CD147, as a host cell co-receptor required for the entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19). The findings reveal that recombinant human basigin does not interact with full-length spike protein or spike receptor-binding domain (RBD) of SARS-CoV-2. The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
It is now well-established that cellular entry of SARS-CoV-2 initiates with the interaction between viral spike protein and host cell angiotensin-converting enzyme 2 (ACE2) receptor. This is followed by proteolytic cleavage and priming of the spike protein by host cell transmembrane serine protease TMPRSS2 and subsequent fusion of the viral envelope with the host cell membrane. Regarding other co-receptors or cofactors of spike glycoprotein, there is preliminary evidence indicating that the spike-basigin interaction plays a vital role in establishing SARS-CoV-2 infection. Based on these observations, clinical trials have been designed to investigate the therapeutic efficacy of anti-basigin monoclonal antibodies against COVID-19.
Basigin, a ubiquitously expressed human transmembrane glycoprotein, plays an essential role in malarial infection. The interaction of basigin with reticulocyte binding protein homolog 5 of malarial parasite Plasmodium falciparum is required for the parasite entry into human erythrocytes.
In the current study, the scientists investigated the involvement of basigin in mediating SARS-CoV-2 host cell entry.
Study design
To explore possible interactions between SARS-CoV-2 and host cell receptors, the scientists generated a panel of recombinant proteins, including full-length spike protein, spike RBD, full-length nucleoprotein, ACE2, and basigin (glycosylated and non-glycosylated). In addition, they used reticulocyte binding protein homolog 5 (RH5) of a malarial parasite as a positive control. The interactions between recombinantly expressed proteins were examined using size exclusion chromatography and surface plasmon resonance.
Important observations
Initially, the scientists conducted a series of size exclusion chromatographic experiments to confirm the quality of recombinant proteins and their interaction patterns. The findings revealed that both full-length spike and spike RBD successfully bind to the human ACE2 receptor. However, no interaction was observed between the RH5 malarial antigen and ACE2. These observations confirm that all recombinant proteins are functionally active and are able to establish expected interactions.
To study the interaction of glycosylated basigin with either full-length spike or spike RBD, they initially conducted a set of experiments using full-length SARS-CoV-2 nucleoprotein and RH5 malarial antigen as negative and positive controls, respectively. While RH5 formed a stable complex with glycosylated basigin, no interaction was observed between full-length nucleoprotein and basigin. Having these confirmations, they next explored the involvement of basigin in the context of SARS-CoV-2 infection and observed that neither full-length spike nor spike RBD interacts with basigin. Moreover, they conducted similar experiments with E-coli-expressed basigin ectodomain to check whether the glycosylation process is affecting the spike-basigin interaction. However, they still could not detect any such interaction.
With the assumption that the spike-basigin complex might not be stable enough to be detected via size exclusion chromatography, they conducted a separate set of experiments using surface plasmon resonance. However, they failed to detect any interaction between spike RBD and glycosylated or non-glycosylated (bacterially-expressed) basigin.
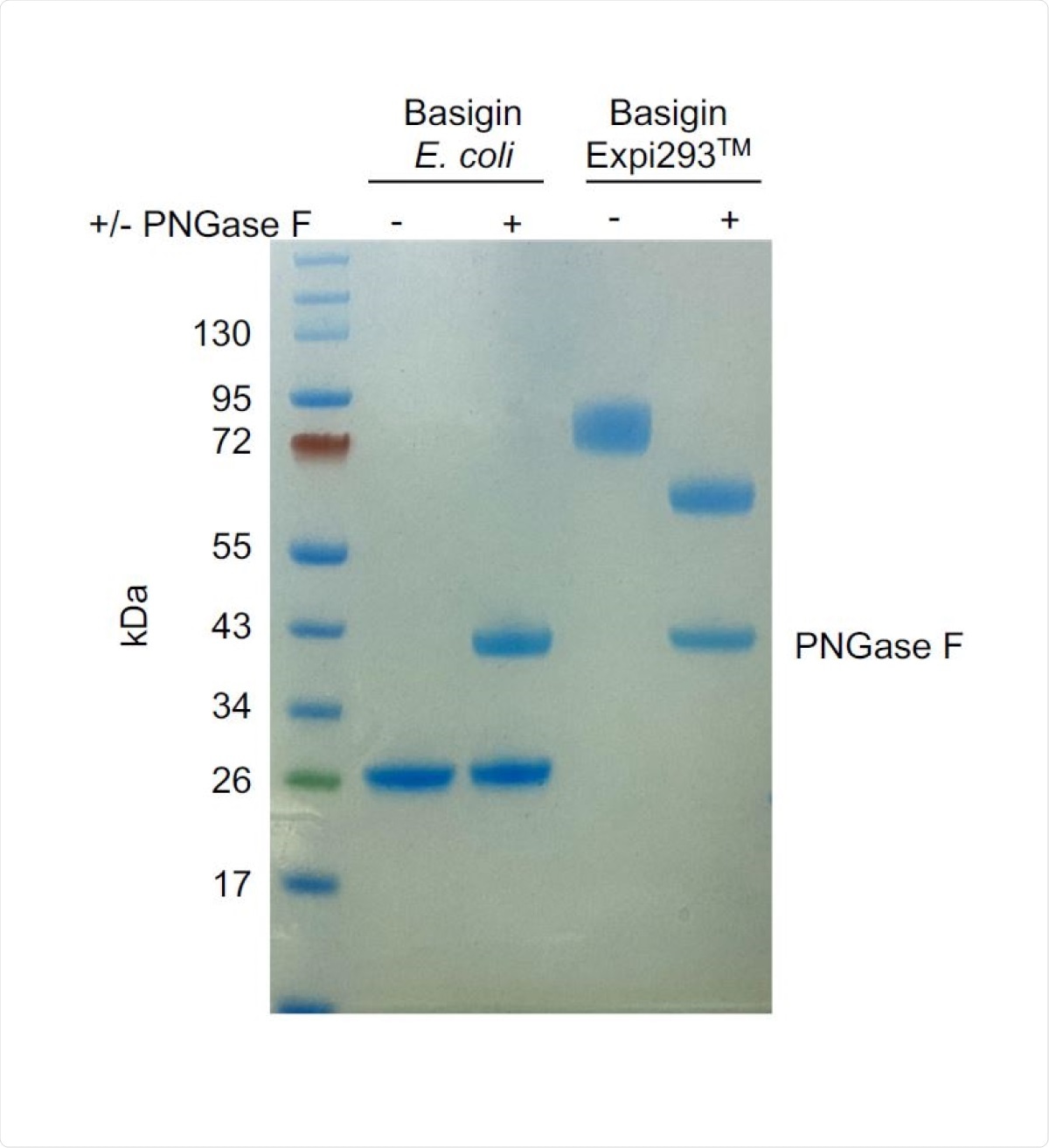
PNGase F digest of E. coli-expressed (non-glycosylated) and Expi293TMexpressed (glycosylated) basigin. The lower molecular weight of glycosylated basigin after PNGase F treatment is consistent with the loss of glycans. The heavier molecular weight of glycosylated basigin treated with PNGase F compared to non-glycosylated basigin can be attributed to the presence of the rat CD4 domains 3+4 (CD4d3+4) solubility tag (33 kDa).
Study significance
The study contradicts previously reported observations on the spike RBD-basigin interaction. It is evident from the current study observations that SARS-CoV-2 does not require basigin as a co-receptor to enter the host cell.
Regarding anti-basigin antibodies, there is preliminary evidence demonstrating the therapeutic benefits of meplazumab, an anti-basigin monoclonal antibody, in managing COVID-19-related pneumonia. However, based on the current findings, the scientists believe that these beneficial effects might be indirectly due to the antibody-mediated reduction of pro-inflammatory responses of basigin. Furthermore, they suggest that the use of basigin as a potential therapeutic target for COVID-19 should be carefully investigated before any clinical implementations.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Robert James Ragotte, David Pulido, Francesca R Donnellan, Giacomo Gorini, Hannah Davies, Juliane Brun, Lloyd DW King, Katherine Skinner, Simon J Draper. 2021. Human basigin (CD147) does not directly interact with SARS-CoV-2 spike glycoprotein. BioRxiv. doi: https://doi.org/10.1101/2021.02.22.432402, https://www.biorxiv.org/content/10.1101/2021.02.22.432402v1
- Peer reviewed and published scientific report.
Ragotte, Robert J., David Pulido, Francesca R. Donnellan, Michelle L. Hill, Giacomo Gorini, Hannah Davies, Juliane Brun, et al. 2021. “Human Basigin (CD147) Does Not Directly Interact with SARS-CoV-2 Spike Glycoprotein.” Edited by Matthew B. Frieman. MSphere 6 (4). https://doi.org/10.1128/msphere.00647-21. https://journals.asm.org/doi/10.1128/mSphere.00647-21.