Genomic surveillance of acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has varied across countries. England's increased coronavirus sequencing helped it uncover the B.1.1.7 variant last fall. In contrast, countries such as the United States have reported little sequencing data.
With an increasing number of variants appearing in various countries, ending the pandemic requires global collaboration in understanding variant mutations.
Mutations in variants found in the United Kingdom, South Africa, Brazil, and California have been shown to increase SARS-CoV-2's survival by increasing the rate of viral transmission or resisting neutralizing antibodies.
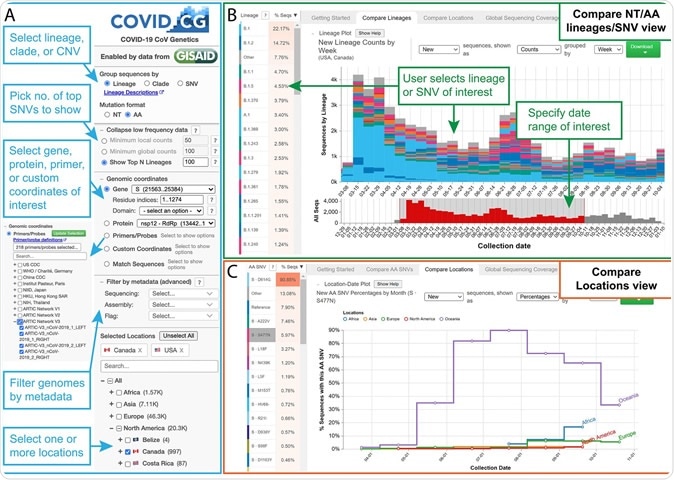
A free public browser called COVID-19 CoV Genetics (COVID-19 CG) was developed to help scientists track and analyze virus mutations by location. This is possible from the collection of viral data available through the GISAID database. Doing so allows COVID-19 CG to track SARS-CoV-2 single-nucleotide variation, lineage, and clades.
Research led by Benjamin E Deverman from the Stanley Center for Psychiatric Research at the Broad Institute of MIT and Harvard provided evidence of COVID-19 CG's effectiveness in tracking SARS-CoV-2 mutations by location.
Understanding mutational changes in the virus are needed to adjust the pandemic response and develop effective treatments.
The authors write:
"Collecting virus genomic data is particularly relevant to regions that are experiencing increases in COVID-19 cases. If only sparse genomic data are sampled, we risk the late detection of SARS CoV-2 variants that exhibit enhanced transmissibility, virulence or resistance against therapeutics or vaccination programs in these pandemic hotspots."
The study, "COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest," was recently published in eLife.
Detecting single nucleotide variation in the spike protein
Single-nucleotide variations in SARS-CoV-2 can change over time. In one case study, the researchers found that an S477N mutation in the spike protein's receptor-binding domain was the dominant coronavirus variant in Australia in December 2020. However, this variant made up only 6% of SARS-CoV-2 genotypes worldwide.
Before July 2020, the RBD N439K mutation was not detected in Ireland but made up 42% of the viral genome there from mid-July to August before declining.
The S477N mutation found in the receptor-binding domain was detected in 1% of Australian sequences before June 2020. However, it comprised 84% of SARS-CoV-2 genetic sequences from June through December.
New SARS-CoV-2 mutation may alter diagnostic methods
The team found several single nucleotide variations that could potentially affect the accuracy of COVID diagnostic primers. They found several variants worldwide containing different C29144T mutations at the very 3' end of the same NIID_2019-nCoV_N_F2 diagnostic primer could change its sensitivity.
Other single nucleotide variations proximal to the 3' ends of primers were also found to potentially change 10 other diagnostic primer pairs used in several other countries.
COVID-19 CG detects new variant found in Australia
The B.1.125 variant with the S477N mutation took over as the dominant strain in Australia between June and September 2020. However, the single nucleotide variations associated with the S477N mutation in Australia differ from those in the United States.
However, this difference may be due to limited genomic surveillance data available in the GISAID database. The researchers also suggest the possibility of sequencing error when there is a singular, sporadic variant.
Because of the reasons above, it is too early to rule out that the variant came from local transmission.
Updates planned for COVID-19 CG database
With variants such as B.1.1.7 becoming the dominant strain in England and soon in the United States, developers of COVID-19 CG plans to upgrade the system with better detection features. The upgrades will help scientists find mutations and variants of concern circulating in local populations more readily.
Given that mutations emerge, disappear, and potentially reappear over time, the research team concludes that countries need to update their genomic surveillance data continually.
"When each country actively contributes to the database of SARS-CoV-2 genomes, this protects against sampling biases that can impact the ability to perform phylogenetic analysis and interpret global SARS-CoV-2 data. Toward this goal that affects all of humanity, we advocate for the increased sequencing of SARS-CoV-2 isolates from patients (and infected animals) around the world, and for these data to be shared in as timely a manner as possible."
Doing so will help vaccine developers update the design and testing of their experimental coronavirus vaccines, antibodies, or other treatments. Scientists can use this data to create hypotheses and test the impact nearby variants have on product-specific interaction interface or antigen.