Last September, researchers at the University of Washington, USA, reported the development of computer-designed small proteins – ‘miniproteins’ – that can target the receptor-binding domain (RBD) on the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic.
In a recent preprint paper published on the bioRxiv* server, the group investigated the miniproteins’ protective capabilities in vivo. The researchers evaluated the miniprotein, LCB1, against SARS-CoV-2-mediated lung disease in human ACE2-expressing transgenic mice.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Here, the researchers used the stringent K18-hACE2 mouse model of SARS-CoV-2 pathogenesis. The K18-hACE2 mouse model recapitulates several aspects of severe COVID-19, such as lung inflammation and reduced pulmonary function.
SARS-CoV-2 belongs to the subfamily Coronavirinae in the family Coronaviridae. These coronaviruses are spherical with a diameter of approximately 80 to 160 mm and are studded with spike glycoproteins. These spike proteins bind to the angiotensin-converting enzyme 2 (ACE2) receptors present on the host cell, binding via the RBD. Following the binding, the spike protein is cleaved by the cell membrane-associated protease, TMPRSS2, which results in membrane fusion enabling the release of the viral RNA genome into the host cell cytoplasm.
Because of the dominant antigenic nature of the spike protein, it has been the primary target of antibody-based countermeasures. To derive an effective countermeasure through the potential loss of binding and the subsequently diminished neutralization, researchers in this study generated a panel of short, 56-amino acid miniproteins.
In the initial research, they set out to design the high-affinity protein minibinders to the SARS-CoV-2 spike receptor-binding domain (RBD) that can compete with the ACE2 binding. They reported high affinity and potent neutralization of authentic virus in cell culture with half-maximal effective concentration (EC50) values < 30 pM.
Our success in designing high-affinity antiviral proteins from scratch is further proof that computational protein design can be used to create promising drug candidates.”
In the current study, they used a stringent model of SARS-CoV-2 disease pathogenesis in human ACE2 (hACE2)-expressing transgenic mice to evaluate the efficacy of one of the miniprotein binders, LCB1, in vivo.
For the in vivo experiments, they evaluated two versions of LCB1: (a) an Fc-modified bivalent form, LCB1-hIgG-Fc9 (LCB1-Fc) that should extend half-life in vivo and engage effector arms of the immune system; and (b) a further optimized, monomeric form of LCB1 lacking an Fc domain, LCB1v1.3.
The researchers reported that the intraperitoneal administration of LCB1-Fc one day pre-or post-SARS-CoV-2 exposure conferred substantial protection. It included an absence of weight loss, and reductions in the viral burden (almost approaching the limit of detection), and inhibition of lung inflammation and pathology. The intranasal delivery of LCB1v1.3 also conferred protection as many as five days before or two days after SARS-CoV-2 inoculation. The intranasal administration is unique and is advantageous over other methods of delivery, as also observed in other studies.
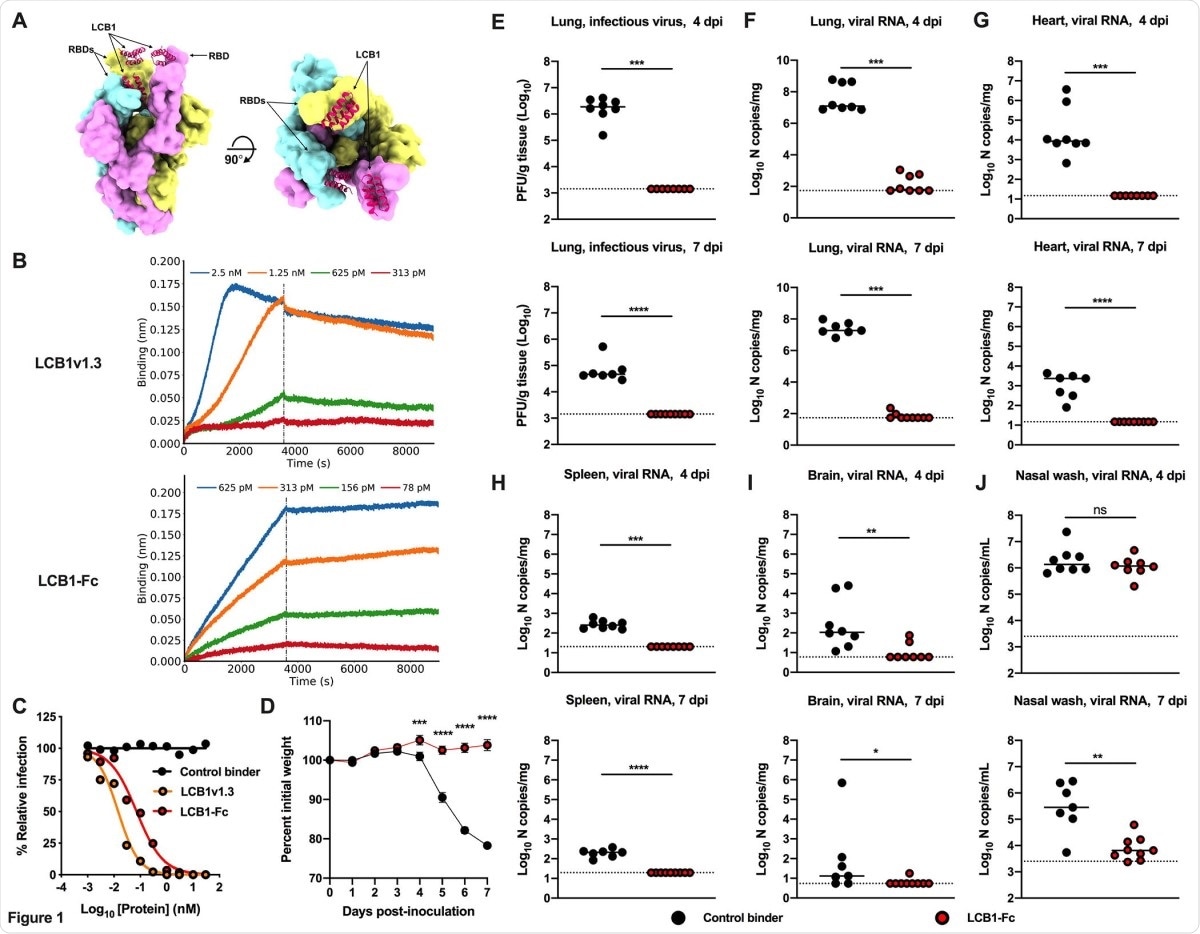
LCB1-Fc prophylaxis protects against SARS-CoV-2 infection. (A) Molecular surface representation of three LCB1v1.3 miniproteins bound to individual protomers of the SARS-CoV-2 spike protein trimer (left: side view; right: top view). (B) Binding curves of purified LCB1v1.3 and LCB1-Fc to SARS-CoV-2 RBD as monitored by biolayer interferometry (one experiment performed in technical duplicate). (C) Neutralization curves of LCB1v1.3, LCB1-Fc, or control binder against a SARS-CoV-2 WA1/2020 isolate (EC50 values: 14.4 pM, 71.8 pM, and >10,000 nM respectively; average of two experiments, each performed in duplicate). (D-J) 7 to 8-week-old female and male K18-hACE2 transgenic mice received 250 µg of LCB1-Fc or control binder by i.p. injection one day prior to i.n. inoculation with 103 PFU of SARS-CoV-2. Tissues were collected at 4 and 7 dpi. (D) Weight change following LCB1-Fc administration (mean ± SEM; n = 8, two experiments: two-way ANOVA with Sidak’s post-test: *** P < 0.001, **** P < 0.0001). (E) Infectious virus measured by plaque assay at 4 or 7 dpi in the lung (n = 8, two experiments: Mann-Whitney test; *** P < 0.001). (F-J) Viral RNA levels at 4 or 7 dpi in the lung, heart, spleen, brain, or nasal wash (n = 8, two experiments: Mann-Whitney test: ns, not significant, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001).
While the dosing experiments revealed that LCB1v1.3 retained efficacy at pharmacologically attainable concentrations and was weakly immunogenic, the researchers have noted that further studies are required to establish the pharmacokinetics. Also, there is a need for efficacy data and further rationale for human clinical trials.
Importantly, the researchers found that the LCB1v1.3 protected animals against the currently emerging B.1.1.7 United Kingdom variant and also a SARS-CoV-2 strain encoding key spike substitutions E484K and N501Y present in both the South Africa (B.1.351) and Brazil (B.1.1.248) variants of concern.
Additional optimization of LCB1-Fc- and LCB1v1.3-RBD binding interactions, through computational design and functional validation, could reduce the effects of variant mutations on neutralizing activity.”
They showed that the LCB1-Fc, an Fc-containing version of the previously reported SARS-CoV-2 RBD binding miniprotein, LCB1, prevented the SARS-CoV-2 infection and disease when administered one day before or after virus inoculation.
In conclusion, these studies have established that these miniproteins – LCB1-Fc and the LCB1v1.3 – are potential candidates for treatment options to prevent or mitigate the SARS-CoV-2 infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
- Longxing Cao, Inna Goreshnik, Brian Coventry, James Brett Case, Lauren Miller, Lisa Kozodoy, Rita E. Chen, Lauren Carter, Alexandra C. Walls, Young-Jun Park, Eva-Maria Strauch, Lance Stewart, Michael S. Diamond, David Veesler, David Baker. (2020) De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 23: doi: https://doi.org/10.1126/science.abd9909, https://science.sciencemag.org/content/370/6515/426
Journal references:
- Preliminary scientific report.
James Brett Case, Rita E. Chen, Longxing Cao, Boaling Ying, Emma S Winkler, Inna Goreshnik, Swathi Shrihari, Natasha M Kafai, Adam L Bailey, Xuping Xie, Pei-Yong Shi, Rashmi Ravichandran, Lauren Carter, Lance Stewart, David Baker, Michael S. Diamond. (2021) Ultrapotent miniproteins targeting the receptor-binding domain protect against SARS-CoV-2 infection and disease in mice. bioRxiv 2021.03.01.433110; doi: https://doi.org/10.1101/2021.03.01.433110, https://www.biorxiv.org/content/10.1101/2021.03.01.433110v1
- Peer reviewed and published scientific report.
Case, James Brett, Rita E. Chen, Longxing Cao, Baoling Ying, Emma S. Winkler, Max Johnson, Inna Goreshnik, et al. 2021. “Ultrapotent Miniproteins Targeting the SARS-CoV-2 Receptor-Binding Domain Protect against Infection and Disease.” Cell Host & Microbe 29 (7): 1151-1161.e5. https://doi.org/10.1016/j.chom.2021.06.008. https://www.cell.com/cell-host-microbe/fulltext/S1931-3128(21)00286-9.