Researchers based in Belgium have developed a new antibody drug that is highly successful at neutralizing coronavirus disease 2019 (COVID-19) in Syrian hamsters. The new biologic was administered to the rodents and was found to be equally successful at neutralizing the original severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain, and also the new mutant variants, such as the South African and UK strains.
Vaccines represent potent tools for combatting diseases, however, they are limited in some regards. Immunity may be short-lived or less effective in old age groups. Limited vaccine availability in many countries, vaccine hesitancy, are other factors of which the impact is currently uncertain.
Passive antibody immunotherapy provides an alternative. Antibodies have long half-lives, are easily and quickly replicable, and, specifically, are capable of being broadly neutralizing. Antibodies with this ability can be more successful within an immune system as they can be effective against multiple mutant variants of a virus, rather than having limited efficacy to one strain.
Nico Callewaert, Xavier Saelens and colleagues have developed a new heavy chain-only antibody, named XVR011, that is equally potent against multiple SARS-CoV-2 variants. Not only that, but it is highly stable, and has “excellent manufacturability.”
Previously, researchers had been able to replicate a prototype antibody, VHH72, that was effective in protecting mice from SARS-CoV-2 infection. In this study, they were able to modify and increase the efficacy of the antibody using computer models. These antibodies were then tested on Syrian hamsters and successfully reduced remnant viral RNA in the lung cavity of the animals.
The team then went on to optimize the antibody molecules, and further tested these antibodies against more virulent strains of the virus in hamsters. This new protein was dubbed XVR011, and was found to be equally potent against the UK and South Africa variants of the virus (B.1.1.7. and B.1.351, respectively). XVR011 is also not reactive with other human proteins, and is specific to viral RNA, supporting the potential use of it for medicinal purposes.
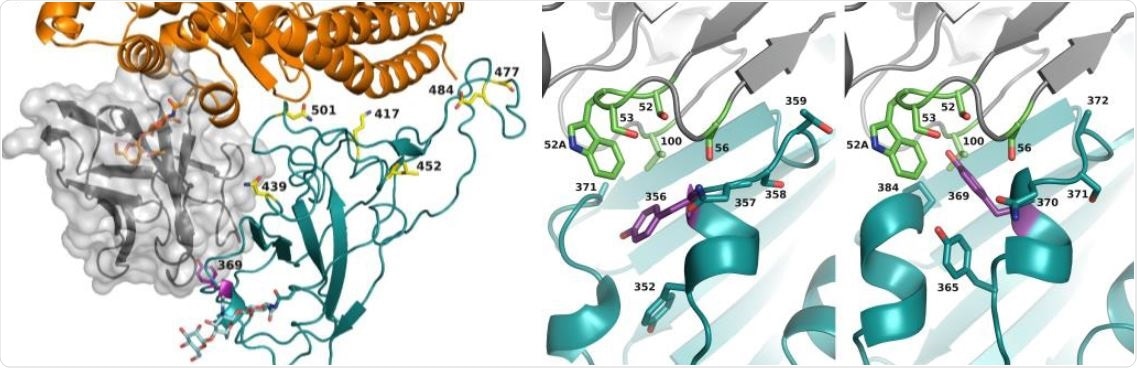
Enhanced affinity and neutralizing activity of computationally predicted VHH72 variant. Left: Composite overlay showing the locations of VHH72 (grey cartoon with the transparent surface, center-left) and ACE-2 (orange cartoon, top) versus SARS-CoV-2 RBD (cyan cartoon, center). Tyr369 of SARS-CoV-2 RBD is indicated and shown as purple sticks. The protein-proximal monosaccharides of the ACE-2 N322 N-glycan (clashing with VHH72) are shown as orange sticks; the RBD N343 N-glycan’s protein-proximal monosaccharides are shown as cyan sticks. The emerging RBD variants at residues K417(->N), N439(->K), L452(->R), S477(->N), E484(->K) and N501(->Y) are indicated and shown as yellow sticks. Right: Comparison of VHH72 (rainbow cartoon) in complex with SARS-CoV-1 RBD (cyan cartoon, pdb-entry 6WAQ chains C and D) with a homology model of VHH72 bound to SARS-CoV-2 RBD (cyan cartoon, model obtained from the I-TASSER server), zoomed-in to the zone near VHH72's S56. VHH72 residues S52, W52a, S53, S56 and V100, SARS-CoV-1 RBD residues Y352, Y356 (purple), N357, S358, T359 and A371, and SARS-CoV-2 RBD residues Y365, Y369 (purple), N370, S371, A372 and P384 are shown as sticks. Figures generated with Pymol (The PyMOL Molecular Graphics System, Open Source Version 2.3. Schrödinger, LLC). RBD Tyr369 assumes a differential preferential conformation between SARS-CoV-1 and SARS-CoV-2, imposed by the P384 in SARS-CoV-2. Accommodating this alteration was the focus of our structure-guided affinity maturation campaign of VHH72.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
“Happy to report on our work… developing a very potent, cross-clade binding, VoC-resistant VHH-Fc antibody drug” tweeted Nico Callewaert. Callewaert is one of the paper's lead authors and a professor at the University of Gent, Belgium.
Such enhanced antibodies may be utilized for longer-term immunity against potential new SARS-CoV-2 variants in the future and could become crucial in protecting populations until they are able to receive a vaccination. Additionally, as this antibody appears to work across multiple variants of SARS-CoV-2, it could be instrumental in slowing the spread of mutant strains which may have previously escaped immunization from current vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Schepens B, et al. Drug development of an affinity enhanced, broadly neutralizing heavy chain-only antibody that restricts SARS-CoV-2 in hamsters. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.08.433449, https://www.biorxiv.org/content/10.1101/2021.03.08.433449v1
- Peer reviewed and published scientific report.
Schepens, Bert, Loes van Schie, Wim Nerinckx, Kenny Roose, Wander Van Breedam, Daria Fijalkowska, Simon Devos, et al. 2021. “An Affinity-Enhanced, Broadly Neutralizing Heavy Chain–Only Antibody Protects against SARS-CoV-2 Infection in Animal Models.” Science Translational Medicine 13 (621). https://doi.org/10.1126/scitranslmed.abi7826. https://www.science.org/doi/10.1126/scitranslmed.abi7826.