New research by German scientists has uncovered another form of coronavirus polymerase. The current polymerase thought to be used for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a viral RNA-dependent RNA polymerase (RdRp) for replication and transcription. It contains a monomeric enzyme with the catalytic subunit nsp12, two copies of subunit nsp8, and one copy of subunit nsp7.
Their findings suggest the two RdRps dimers in the alternative structure help with template switching when the virus is preparing sub-genomic RNAs for transcription.
The study “Dimeric form of SARS-CoV-2 polymerase” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
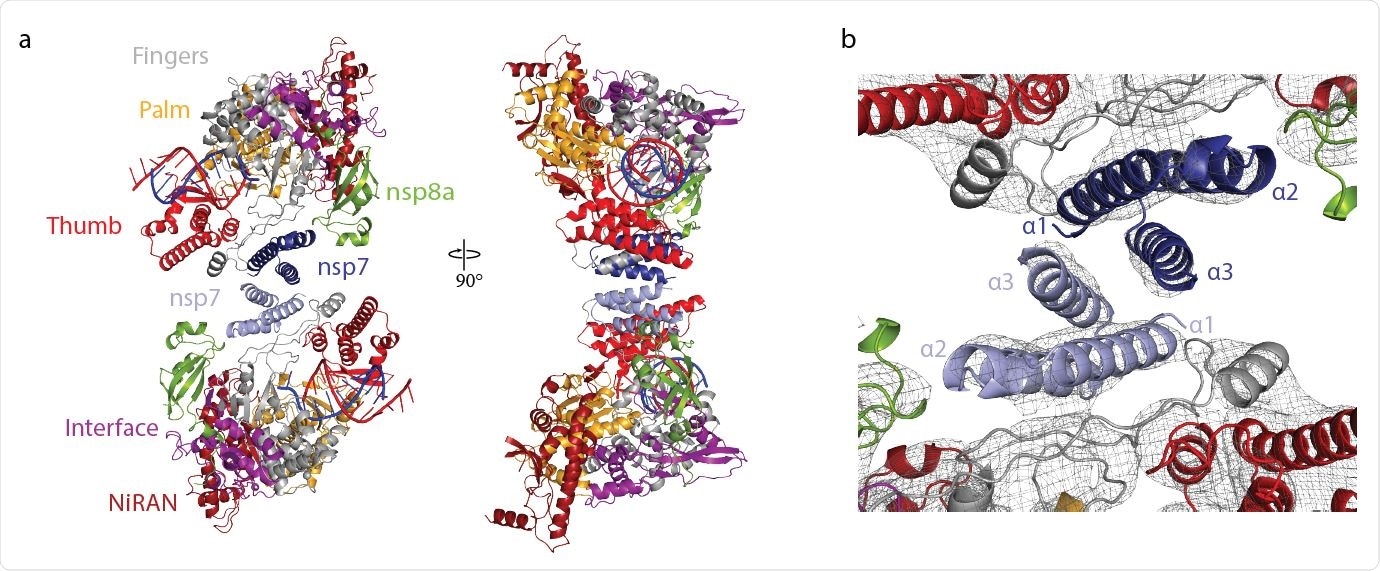
Structure of antiparallel RdRp dimer. a Two views of a ribbon model of the antiparallel RdRp-RNA dimer. Color code for nsp7, nsp8, nsp12 domains (NiRAN, interface, fingers, palm and thumb), RNA template (blue) and RNA product (red) is used throughout. Nsp7 subunits in the two RdRp monomers are colored slightly differently for the two monomers (dark and light blue, respectively). Views are related by a 90° rotation around the vertical axis. b Close-up view of nsp7-nsp7 dimerization interface. View is as in the left structure of panel a. The final cryo-EM density is shown as a black mesh.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
How they did it
The researchers wrote a script to search for higher-order Rd-Rp assemblies in cryo-EM data. They calculated nearest-neighbor (NN) distances and relative orientations of neighboring RdRp enzymes, where many were at a NN distance of 80 Å and a relative orientation of 180°. The research team suggests this provides more evidence of a structurally defined RdRp dimer.
About 27,473 dimer particles showed a strong RNA signal in 2D and had a 3D reconstruction at an overall resolution of 5.5 Å. When they obtained the density with the two RdRp-complexes, they found an antiparallel arrangement with RNA exiting opposite sides of the dimeric particle. The resolution increased to 4.8 Å with 3D reconstruction.
“The reconstruction unambiguously showed that both RdRps lacked one copy of nsp8, and thus each enzyme was comprised of only one copy of nsp12, nsp8, and nsp7,” wrote the team. They note the final structure was created through rigid-body fitting.
The researchers observed the two polymerases interacted by their nsp7 subunits with contact between the nsp7 helices α1 and α3. The interaction with the nsp7-nsp7 dimer interface occurs after nsp8b dissociates, freeing the dimerization region of nsp7. The team notes the nsp7-nsp7 formation corresponds to an nsp7-nsp8 heterotetramer and how it dissociates into a stable nsp7-nsp8 heterodimer that may bind to nsp12.
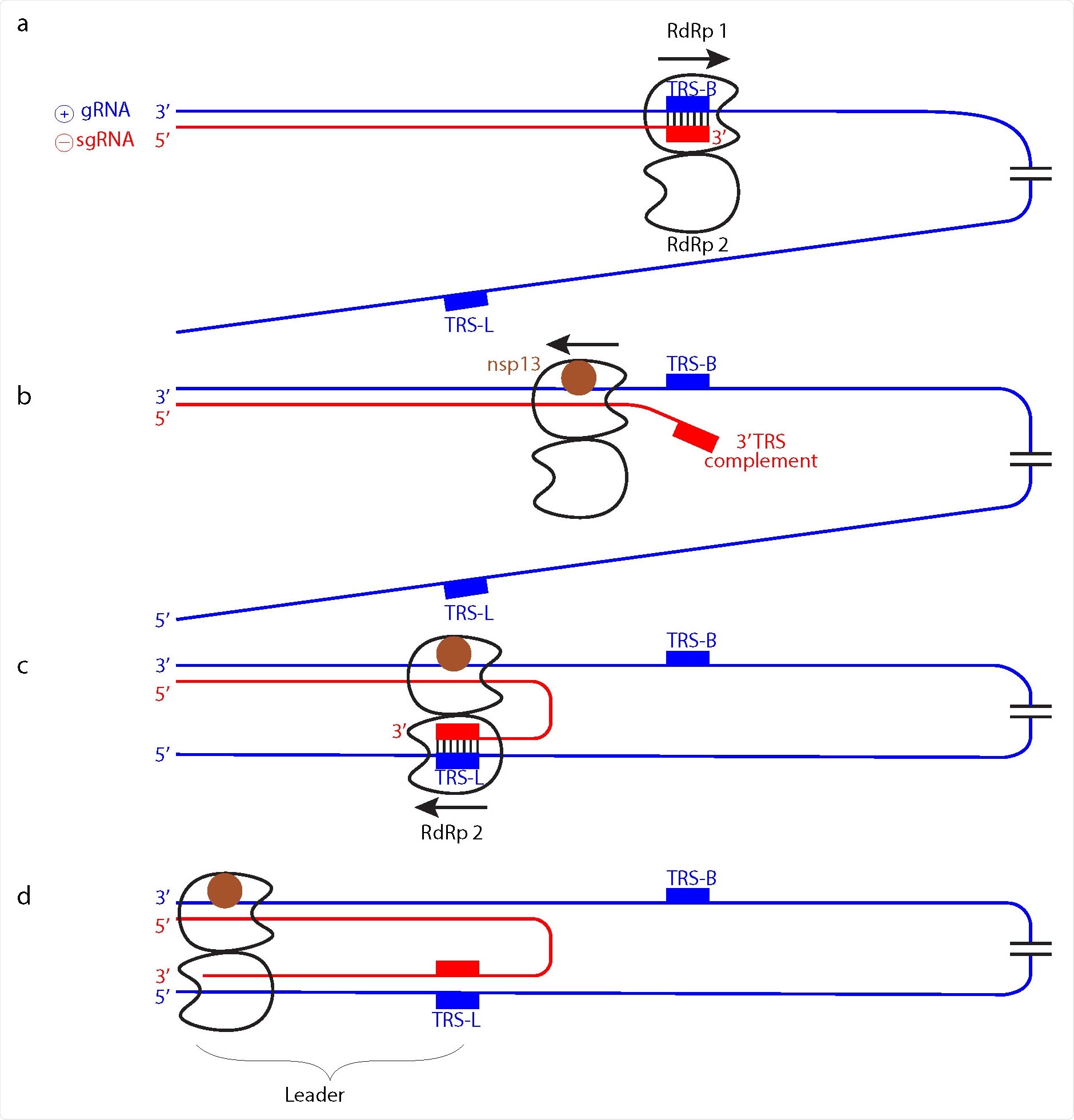
Model of subgenomic RNA production for transcription. a Genomic positive-strand (⊕) RNA (gRNA) is used as template to produce the 5’ region of negative-strand(⊖) sgRNA until TRS-B is reached by the RdRp monomer 1. b Backtracking is mediated by nsp13 helicase and exposes the newly synthesized, complementary TRS sequence. c The complementary sequence in sgRNA can pair with the downstream TRS-L in gRNA and is then loaded into RdRp monomer 2. d RdRp 2 then completes ⊖ sgRNA synthesis while RdRp 1 backtracks further.
There is a possibility of mutation in the SARS-CoV-2 genome that may influence the relative stability of the RdRp dimer. Previous work has found the nsp12 mutation P323L coevolving with the D614G mutation and is mainly found in severely affected patients. In addition, the nsp7 mutation S25L may be involved in destabilizing the binding of nsp7 to nsp8a22 and the nsp7 mutation L71F. This interaction has been associated with severe COVID-19 and could further destabilize the binding of the nsp7 C-terminal region to nsp8b.
However, the team notes that the catalytic sites and the RNA duplexes were not involved in RdRp dimer formation. This suggests the two RdRp enzymes may keep working in the dimer structure, continuing RNA-dependent processes — although the researchers could not test this because of purification issues.
Based on the findings, they suggest the function of the RdRp dimers ties in with subgenomic RNA synthesis, which is used as a template to make viral mRNAs.
“In the model, one RdRp of the dimer (RdRp 1) synthesizes sgRNA from the 3’ end of the gRNA template until it reaches a TRS in the template body (TRS-B). Due to the lack of one nsp8 subunit, the dimeric RdRp is predicted to have lower processivity than monomeric RdRp10,14, and this may facilitate TRS recognition.”
Additionally, it is the RNA, not the RdRp, that switches to a second RdRp position. Other movements include the prediction of only one RdRp in the dimer backtracking in the strand. Specifically, it may be RdRp 1 backpedaling to the 3’ end of the template while RdRp2 moves forward in the opposite direction towards the 5’ end of the template.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Jochheim FA, et al. Dimeric form of SARS-CoV-2 polymerase. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.23.436644, https://www.biorxiv.org/content/10.1101/2021.03.23.436644v1
- Peer reviewed and published scientific report.
Jochheim, Florian A., Dimitry Tegunov, Hauke S. Hillen, Jana Schmitzová, Goran Kokic, Christian Dienemann, and Patrick Cramer. 2021. “The Structure of a Dimeric Form of SARS-CoV-2 Polymerase.” Communications Biology 4 (1). https://doi.org/10.1038/s42003-021-02529-9. https://www.nature.com/articles/s42003-021-02529-9.