The effect of different neutralizing antibodies, both vaccine-elicited and from convalescent sera, suggests new variants are emerging that can evade the human immune response.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) binds to host cells via its spike protein, which has two parts, the S1 and S2 subunits. The S1 subunit has the receptor-binding domain (RBD) and the N-terminal domain (NTD). The RBD binds to the host angiotensin-converting enzyme 2 (ACE2) to infect host cells.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Neutralizing antibodies are produced against both the RBD and NTD by infected and vaccinated individuals. Some RBD-specific monoclonal antibodies are currently under clinical trials or approved for use to treat COVID-19 patients.
The virus's continued spread has led to it evolving rapidly, with several variants emerging in different parts of the globe. The variants B.1.1.7, B.1.351, and the P.1 strains have several mutations in the spike portion and other regions of the viral genome. Some of these mutations have reduced the neutralizing potency of monoclonal antibodies, convalescent sera, and some vaccines.
Another variant, B.1.427/B.1.429 emerged in California in May 2020 and has now spread to several other countries. The B.1.427/B.1.429 variant has more than 50% prevalence in California since February 2021. It has mutations S13I, W152C in the NTD, and L452R mutation in the RBD. This has been listed as a variant of concern (VOC) by the United States Centers for Disease Control (CDC) because of its rapid spread.
In a study published on the bioRxiv* preprint server, researchers report the effect of neutralizing antibodies from recovered patients or mRNA vaccinated individuals on neutralizing this variant.
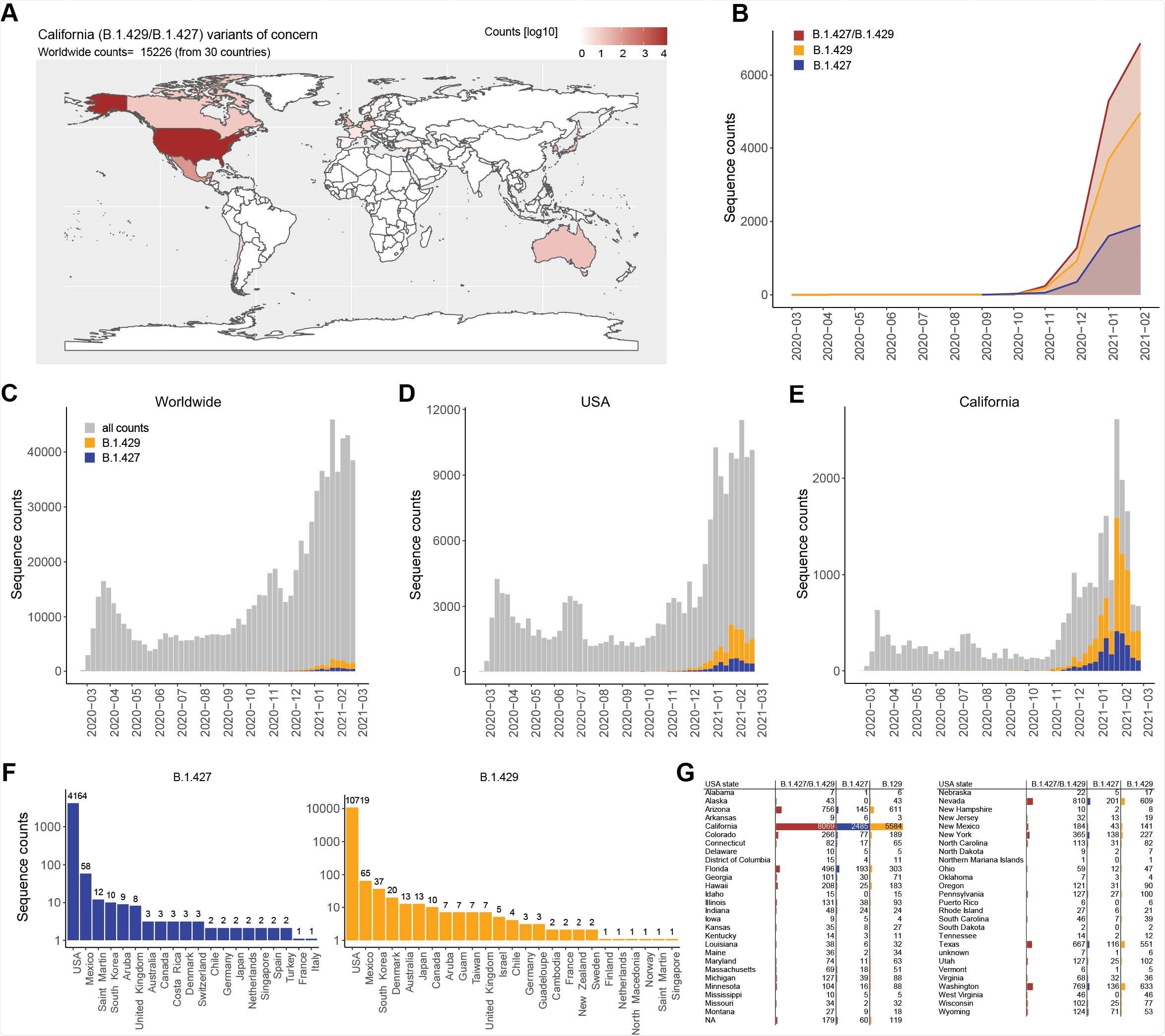
Geographic distribution and evolution of prevalence over time of the SARS-CoV- 2 B.1.427/B.1.429 VOC. (A) World map showing the geographic distribution and sequence counts of B.1.427/B.1.429 VOC as of March 26, 2021. (B) Cumulative and individual B.1.427/B.1.429 VOC sequence counts by month. (C-E). Total number of SARS-CoV-2 (grey) and B.1.427/B.1.429 VOC (blue/orange) sequences deposited on a monthly basis worldwide (C), in the US (D) and in California (E). (F and G) Total number of B.1.427 and B.1.429 sequences deposited by country (F) and by US states (G) as of March 26, 2021.
Testing antibody neutralization of B.1.427/B.1.429 strain
Using a lentiviral system, the researchers saw that the neutralizing activity of the antibodies elicited by the Moderna and the Pfizer/BioNtech vaccines was reduced about 3-fold and 4-fold, respectively, similar to that of the B.1.351 strain and slightly higher than the neutralizing activity toward the B.1.1.7 strain and the P.1 strain.
Sera obtained from people who recovered from COVID-19 after being infected from the original Wuhan-1 strain showed about a 5-fold reduction in the neutralization of the B.1.427/B.1.429 strain compared to the D614G strain, comparable to that seen for other mutant strains. This indicates antibody responses elicited by vaccination are of a little higher quality than those from natural infection.
Sera obtained from people infected with B.1.1.7 showed almost undetectable neutralizing activity against the D614G variant and other VOCs, including B.1.427/B.1.429. This suggests infection with the B.1.1.7 variant, the dominant lineage in Europe and some other countries, produce low levels of cross-neutralizing antibodies against other variants.
Next, the team compared the neutralizing activity of several RBD- and NTD-specific monoclonal antibodies to the D614G strain and the B.1.427/B.1.429 spike mutations using a pseudo-type virus system.
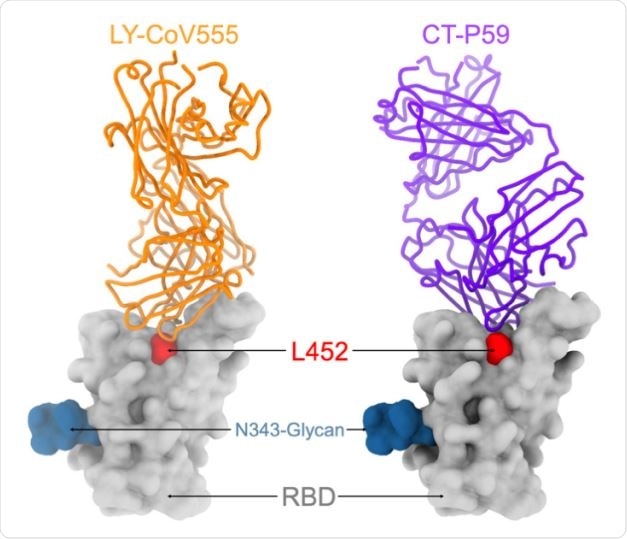
Surface representation of the SARS-CoV-2 RBD (grey) bound to the bamlanivimab (LY-CoV555, orange, PDB 7CM4) and regdanvimab (CT-P59, purple, PDB 7KMG) Fab fragments shown as ribbons. The L452 side chain is shown as red spheres to indicate its central location within the epitopes of these two mAbs. The N343 glycan is rendered as blue spheres.
Out of 35 antibodies they tested, they found 14 had reduced neutralizing potency. The effect of the casirivimab/imdevimab antibody cocktail that received emergency use authorization in the United States did not change because of the L452R mutation.
Ten monoclonal antibodies specific to the RBD showed 10-fold reduced neutralization of B.1.427/B.1.429 and had poor binding to L452R RBD mutant. The S31I and W152C mutations removed the neutralization effect of NTD-specific monoclonal antibodies.
Strong selective pressure for mutations
Because the S31I mutation is present in the signal peptide and is thought to shift the signal peptide cleavage site, it is likely the mutation affects the integrity of the NTD antigenic site, compromising the N-terminus. Along with the W152C mutation, this is potentially responsible for the B.1.427/B.1.429 variant escaping from NTD-specific antibodies.
The immune evasion of L452R mutation in the B.1.427/B.1.429 spike protein is similar to previous findings of this mutation, even before the identification of the B.1.427/B.1.429 variant. The acquisition of this mutation across many lineages and continents suggests positive selection resulting from the selective pressure of RBD-specific neutralizing antibodies.
Unlike the antibodies targeting the RBD, NTD-specific antibodies target only one antigenic supersite. Hence, the SARS-CoV-2 NTD undergoes fast antigenic drift and has a lot of mutations, suggesting it is under strong selective pressure from the host immune response. Understanding the various mechanisms of immune evasion of different virus variants could help develop effective strategies to combat the pandemic.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
McCallum, M. et al. (2021) SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv, https://doi.org/10.1101/2021.03.31.437925, https://www.biorxiv.org/content/10.1101/2021.03.31.437925v1
- Peer reviewed and published scientific report.
McCallum, Matthew, Jessica Bassi, Anna De Marco, Alex Chen, Alexandra C. Walls, Julia Di Iulio, M. Alejandra Tortorici, et al. 2021. “SARS-CoV-2 Immune Evasion by the B.1.427/B.1.429 Variant of Concern.” Science, July. https://doi.org/10.1126/science.abi7994. https://www.science.org/doi/10.1126/science.abi7994.