Using three classes of antibodies, researchers modeled their binding to the SARS-CoV-2 virus spike protein. They found binding to key residues in current mutations changes how the spike protein moves and can lead to binding resistance.
Some antibodies can bind to different parts of the spike protein simultaneously, providing a more potent neutralizing capacity. Class I antibodies have a considerable overlap with the angiotensin-converting enzyme 2 (ACE2) receptor and bind when the receptor-binding domain (RBD) is in the open or up conformation.
Class II antibodies have only a partial overlap with the ACE2 epitopes. They can bind to the RBD both in the up and down conformation and have more diverse neutralization mechanisms. Class III antibodies bind to a more inaccessible RBD site that requires at least two RBDs to be in the up conformation.
This suggests a cocktail of antibodies will work better at neutralizing the virus than a single antibody alone. In a study published in the bioRxiv* preprint server, researchers report the structure of binding complexes of the different classes of antibodies to the spike RBD using computational modeling, which may also help explain how the virus generates escape mutations.
Characterizing binding of different classes of antibodies
The team used the structures of the RBD with B38, a class I antibody; P2B-2F6, a class II antibody; and EY6A and S304, class III antibodies. They used atomistic molecular dynamics simulations and mutational scanning to determine the changes in protein stability.
The analysis showed that the most extensive changes induced by binding were for the B38 antibody around the K417 site. In the RBD-B38 complex, the positions K417 and N501 were stabilized, but the residues around E484 were more flexible. The binding characteristics for class II antibodies were similar, with a little more flexibility than the RBD-B38.Class III antibodies binding showed stabilizing of the residues and the RBD core became more rigid.
Mutations around the E484 position can make the virus resistant to antibody neutralization. However, class III antibodies are not much affected by this. Although EY6A does not create any structural hindrance to ACE2 binding, because of its ability to change the stability of the receptor-binding motif (RBM), it may hinder ACE2 binding.
When an antibody binds to the spike protein, it can give rise to a change in its conformation because of the binding, called an allosteric response. Analysis revealed binding to the flexible regions near K417 and N501 by Class I antibodies may give rise to such as response. Binding at Y449 and E484 by Class II antibodies modulates allosteric changes.
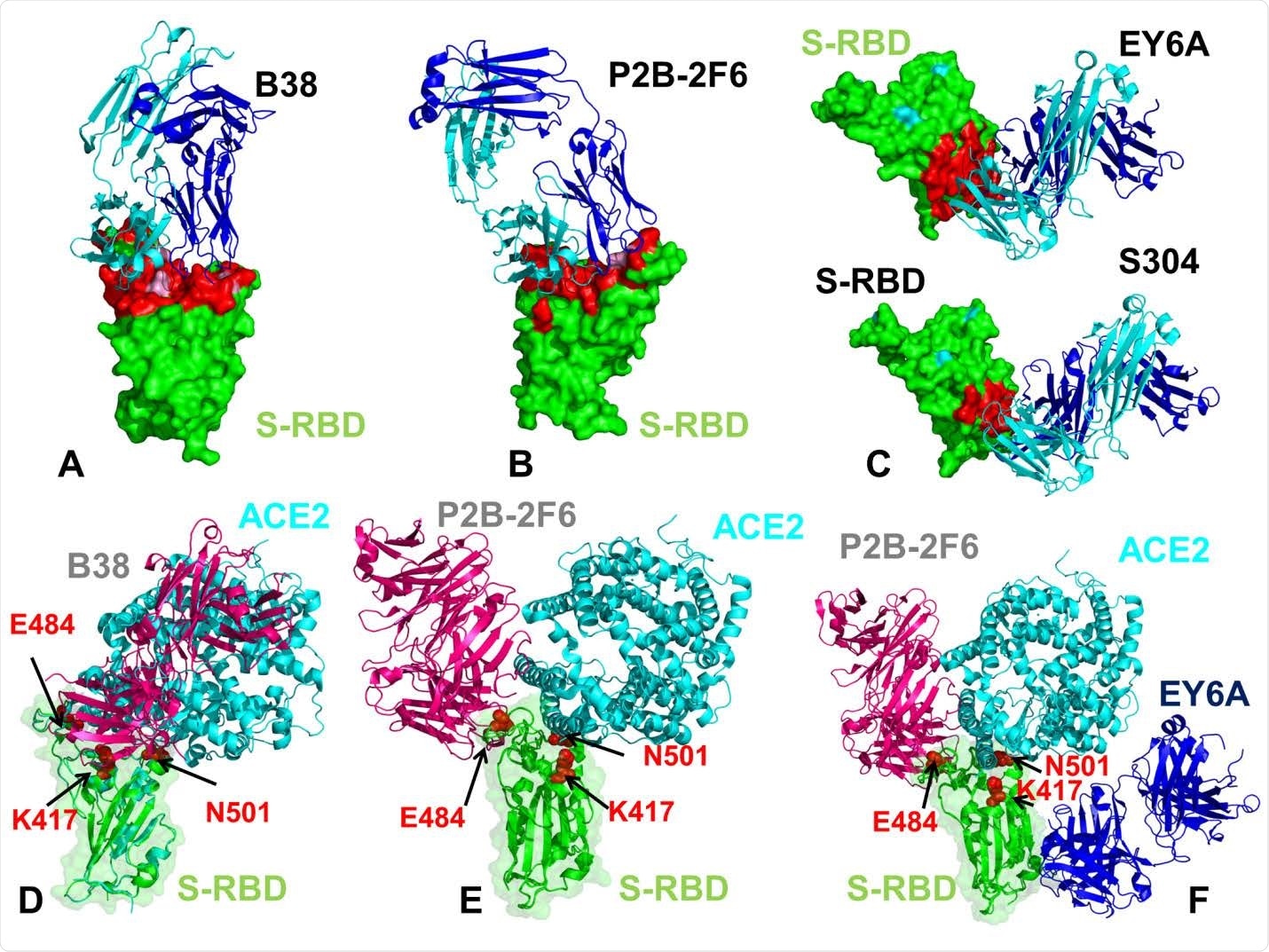
The structures of the SARS-CoV-2 S-RBD complexes with a panel of antibodies used in this study. (A) The structure of the SARS-CoV-2 S-RBD complex with B38 (pdb id 7BZ5). (B) The structure of the SARS-CoV-2 S-RBD complex with P2B-2F6 (pdb id 7BWJ). (C) The structures of the SARS-CoV-2 S-RBD complex with EY6A (pdb id 6ZER,6ZCZ)36 and S304 antibodies. S-RBD is shown in green surface and the binding epitopes are colored in red. The antibodies are shown in ribbons with heavy chain colored in blue and light chain in cyan. (D) Structural superposition of the B38 antibody ( in pink ribbons) and ACE2 host receptor (in cyan ribbons) bound to S-RBD ( in green ribbons and surface with reduced transparency). (E) Structural superposition of the P2B-2F6 antibody ( in pink ribbons) and ACE2 host receptor (in cyan ribbons) bound to S-RBD (in green ribbons). (F) Structural superposition of the P2B-2F6 antibody (in pink ribbons), ACE2 host receptor (in cyan ribbons) and EY6A (in blue ribbons) bound to S-RBD (in green ribbons). The positions are sites K417, E484 and N501 subjected to circulating mutational variants are shown in red spheres and annotated.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Dynamics of antibody-spike complexes
The team’s analysis revealed that the currently circulating mutations are present on sites that are undergoing movements along with slow-mode motions of the spike protein. But, none of the mutation sites, K417, E484, and N501 are on the hinge positions of the unbound RBD. Targeting these hinge regions may change the slow-mode movement of the spike and change functional characteristics.
The class I antibody, B38, which binds to the K417 region, altered the protein mobility, suggesting binding can change the location of hinge sites. For class II antibodies, G446, Y449, and E484 positions are key for functional motions of the proteins, and mutations on these positions could change P2B-2F6 antibody binding.
Class III antibodies showed a different effect upon binding. They change conformations slightly at the ACE2 binding site rather than dramatic large-scale motions. The model suggests there could be interactions between the bound antibodies and ACE2.
Mutational sensitivity analysis showed that class I antibodies have several binding sites and share binding sites with ACE2. Class II antibodies bind to a smaller region on the spike protein. Both the Class III antibodies tested bind to the same sites.
Energy analysis indicated that a mutation at the K417 site led to a significant destabilization of the spike protein. Mutations at N501 were also detrimental to antibody binding. Thus, the circulating mutations K417N and N501Y could lead to a loss of B38 antibody binding.
Although mutations at the E484 site led to very small changes, all mutations at this site led to a significant loss in antibody binding, including the circulating strain E484K in the B.1.351 variant. This mutation could lead to Class I and Class II antibody escape. Thus, the spike protein may use the allosteric effects caused by antibody binding to generate escape mutants that can change antibody binding but not affect the spike functioning.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources