A team of international scientists recently demonstrated that antibodies generated in response to the DNA vaccine INO-4800 can effectively neutralize some of the recently emerged variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), such as G614, 501Y.V1, and 501Y.V2. The study is currently available on the bioRxiv* preprint server.
The coronavirus disease 2019 (COVID-19) vaccines currently rolling out worldwide have mostly been developed against the original strain of SARS-CoV-2 (D614). With the recent emergence of novel variants of concern (VOC), the globe faces a new challenge of the effectiveness of existing vaccines on new viral variants. In this context, a growing pool of evidence has suggested that spike mutations present in these VOCs can reduce the neutralizing efficacy of antibodies developed in response to COVID-19 vaccines or natural infections with previously circulating viral variants.
In the current study, the scientists have investigated whether antibodies developed in response to a leading DNA vaccine INO-4800 can effectively neutralize the UK (501Y.V1), South African (501Y.V2), and G614 variants of SARS-CoV-2. The vaccine, which has been developed by Inovio Pharmaceuticals, San Diego, USA, contains the spike protein-coding sequence of the original Wuhan strain of SARS-CoV-2 as an immunogen.
Previous studies have shown that antibodies induced by mRNA-based COVID-19 vaccines are less effective in neutralizing both the UK and South African VOCs. Similarly, serum samples obtained from COVID-19 patients in South Africa have shown significantly reduced neutralizing efficiency against the South African variant.
Study design
The INO-4800 vaccine examined in the study has good safety and efficacy profiles in humans, as observed in clinical trials. Moreover, the vaccine can be stored at room temperature, which is an added benefit in relation to vaccine transport to remote places.
To induce anti-SARS-CoV-2 antibody responses, a total of 8 ferrets were immunized intramuscularly with two doses of INO-4800 vaccine at an interval of 28 days. The serum samples were collected on day 35 and day 42 post-vaccination to ensure sufficient seroconversion.
Using Vero cells, cytopathic effect (CPE) inhibition assays were carried out to examine the virus neutralization efficacy of immunized ferret sera.
Important observations
The scientists estimated the average neutralization titers for each serum sample against the tested viral variants. The findings indicated that the neutralization titers against G614 and 501Y.V1 were comparable. However, the titers against the 501Y.V2 variant were significantly lower (4-fold) than that against the other two variants.
To conduct molecular modeling experiments, the scientists developed fully glycosylated spike protein models of three viral variants. In addition, they included one angiotensin-converting enzyme 2 (ACE2) protein bound to the spike receptor-binding domain (RBD). Finally, they simulated these models in an aqueous solution using specialized software.
The findings of molecular modeling experiments revealed the structural alterations in the 501Y.V2 variant were more pronounced than the 501Y.V1 variant, which further justify the lower neutralization titers observed against 501Y.V2. Similarly, more extensive alterations were observed in the N-terminal domain (NTD) of the 501Y.V2 variant, with a group of mutations including L18F, D80A, D215G, and a 242-244 deletion.
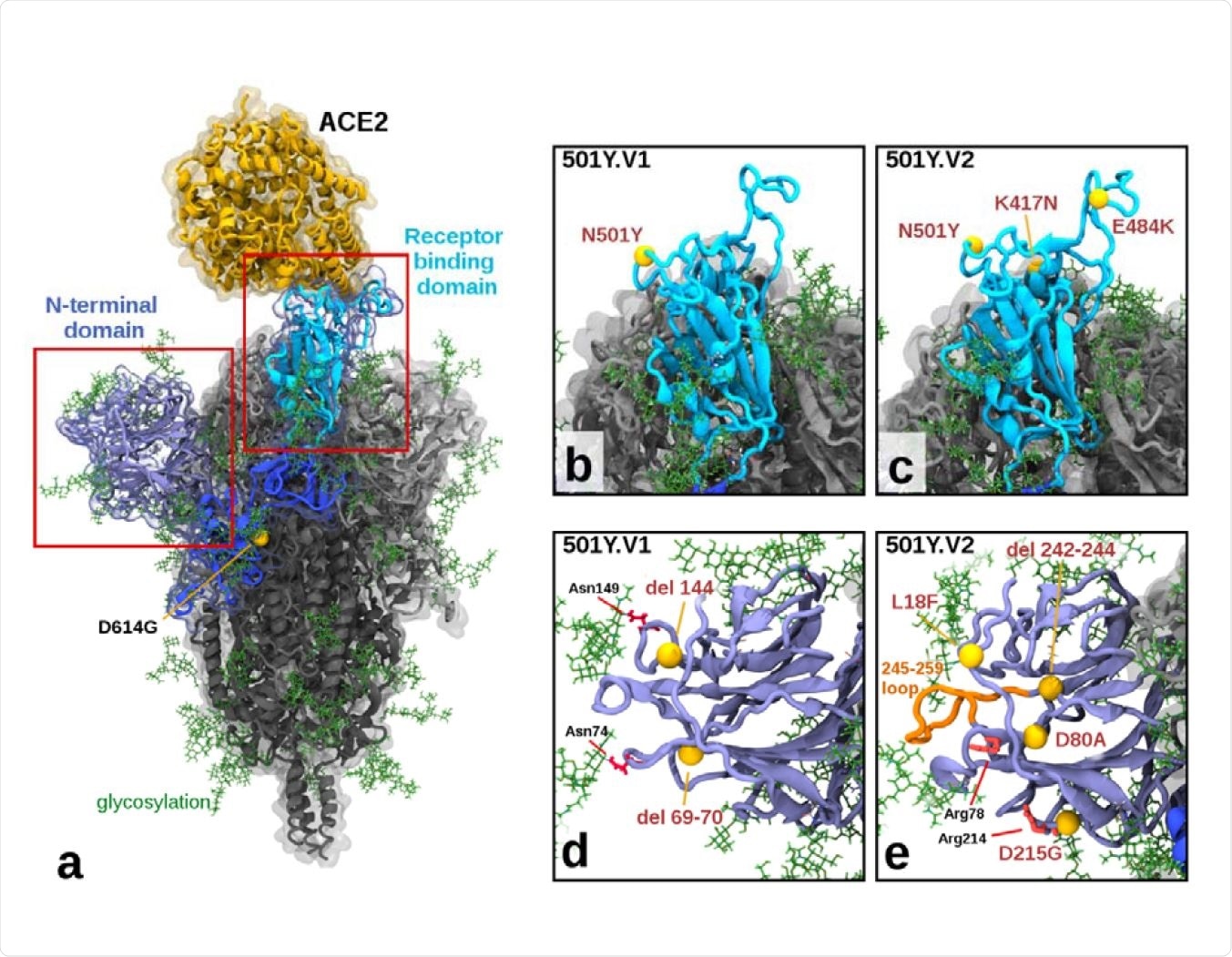
Illustration of Mutations in 501Y.V1 and 501Y.V2 Relative to the Ancestral Form a) Structure of the glycosylated SARS CoV2 Spike protein highlighting a S1 monomer (in blues) and relative positions of the N terminal domain (NTD) and the receptor-binding domain (RBD) and the bound ACE2 receptor (in yellow). The position of D614G (common to all tested variants) is also highlighted. b) & c) Side by side comparison of 501Y.V1 and 501Y.V2 variants in the RBD, showing the V2 variant having additional K417N and E484K mutations. d) & e) Side by side comparisons of 501Y.V1 and 501Y.V2 variants in the NTD, showing relative locations of mutations and deletions. (Model files available in Supplementary Materials).
The N501Y mutation present in the spike RBD of the 501Y.V1 variant has been found to increase its binding affinity for the host cell ACE2 receptor. In addition to the N501Y mutation, the spike RBD of the 501Y.V2 variant contains two other mutations K417N and E484K. Collectively, these mutations have been found to increase the infectivity and immune evasion ability of the virus.
In addition, the mutations observed in the NTD of the 501Y.V2 variant are expected to influence the antigenic presentation and glycosylation arrangements of the spike protein. Taken together, the structural alterations observed in the 501Y.V2 variant justify its potency to reduce the neutralizing ability of INO-4800-induced antibodies.
Study significance
The study reveals that antibodies induced by the DNA vaccine INO-4800 can effectively neutralize the G614 and 501Y.V1 variants of SARS-CoV-2. However, the vaccine is not equally effective against the 501Y.V2 variant.
The 501Y.V2 neutralizing ability of the INO-4800 vaccine is comparable to the mRNA-based COVID-19 vaccines developed by BioNTech/Pfizer and Moderna. In contrast, the adenovirus-based vaccine developed by Oxford/AstraZeneca has shown significantly lower efficacy in neutralizing 501Y.V2.
Given these observations, the scientists believe that nucleic acid-based COVID-19 vaccines are more effective than other vaccine types in neutralizing novel VOCs.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources