Increasing genomic surveillance and profiling high-frequency clusters of missense mutations would help develop therapeutics and vaccine targeting viral variants.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Collecting data to form the CovSimulator software
The researchers used the first known SARS-CoV-2 genome collected from Wuhan, China, in December 2019 to serve as the CovSimulator — a model simulating the evolution of the genome.
When looking at approximately 1 million SARS-CoV-2 genomes up to March 31, 2021, a custom database was created and profiled 815,402 genomes. About 70% of mutations from genomes were single-nucleotide substitutions, including C>T or G>T substitutions.
Rise of hyper-mutated viral variants
The researchers took about 100 genome samples from six continents — excluding Antarctica — to look at mutational differences. They found the rate of mutations varied in viral samples circulating in Asia, Europe, Oceania, and South America since October 2020.
The emergence of variants after October 2020 appeared to be attributed to multiple missense mutations with little viral genome divergence. However, the researchers note the missense divergence happened before the emergence of new viral lineages in North America.
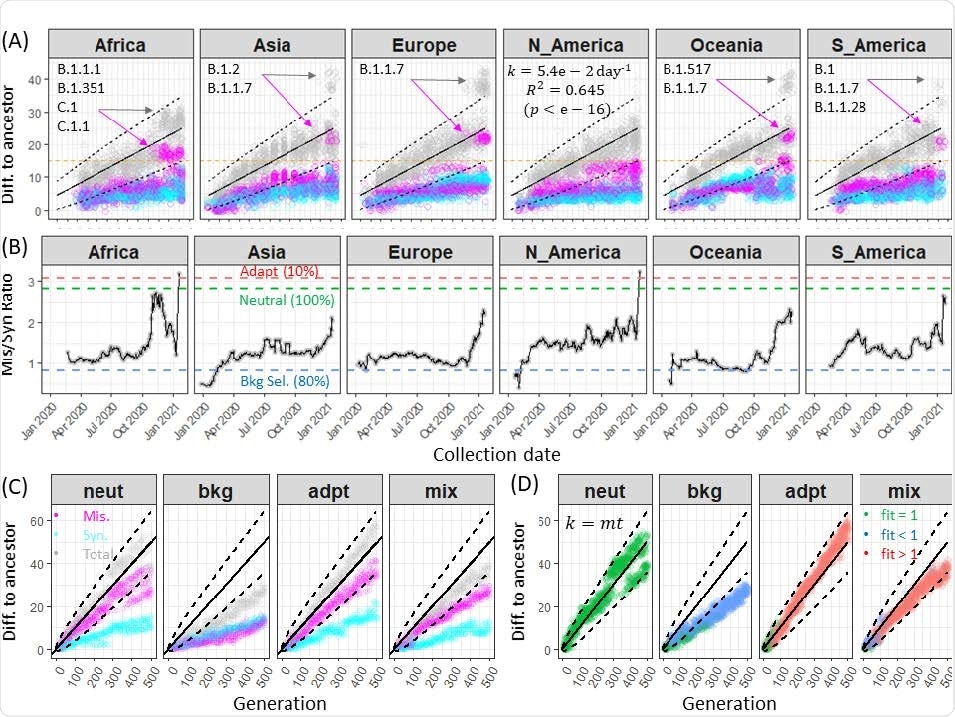
Rates of synonymous and nonsynonymous divergence of SARS-CoV-2 genomes
Driving forces behind SARS-CoV-2 mutations
Variants with beneficial mutations that allowed it to survive and spread overtook the original SARS-CoV-2 strain as the dominant viral lineage.
While mutations were expected to occur with the virus, the researchers questioned whether SARS-CoV-2 accelerated its mutation rate because of other outside forces.
Simulation of the SARS-CoV-2 genome showed high genomic diversity in the six continents during 2020. “Clearly, the global viral populations are far from reaching an equilibrium level of genomic diversity as the virus has spread within and across continents, mirroring the failures in local and global outbreak control. Furthermore, the increasing genomic diversity may be a reflection of increasing admixture of viral subpopulations distributed across the continents,” wrote the team.
Another potential factor towards viral genomic diversity was the relaxation of selective constraints and adaptive mutations.
Mixed SARS-CoV-2 evolution
Despite most missense mutations being deleterious, adaptive mutations and strong genome-wide linkage disequilibrium appeared to be the main drivers behind genomic variability, suggesting SARS-CoV-2 underwent a mixed genome evolution.
When sampling 20 SARS-CoV-2 genomes, there was a shortened time since the most recent common ancestor. There were also 11 out of 19 adaptive mutations that dominated the viral population.
“Critically, it is clear from the Muller diagrams that within each “genotype” (e.g., G1, G2, G3, G8, and G9), at least one genetic change was the driver adaptive mutation.”
Characterizing adaptive mutations
The mixed SARS-CoV-2 genomic evolution model showed that adaptive mutations were frequent among single-nucleotide variations. About 10.9% of missense mutations were adaptive mutations in a population sample of genomes. In 31 missense mutations, 45% of adaptive mutations reached a frequency of 0.5% or higher.
The rate of adaptive mutations among missense mutations increased over time.
Twenty genomes from the previous viral generation showed fixed missense mutations were comprised of 70% adaptive mutations, 20% deleterious mutations, and 10% neutral mutations. At least one — G1, G2, G3, G8, and G9 — were adaptive drivers for mutation in clusters of mutations.
The researchers next looked at the mutation rate over time for each continent. There were approximately 52 missense mutations on the SARS-CoV-2 spike protein with more than 5% frequency in at least a month.
These mutations, including the D614G substitution, appeared to form in clusters that were distributed worldwide. Other spike protein mutations that make up several variants of concern, such as B.1.351, were also identified.
But not all variants and their associated mutations experienced a global transmission. When researchers looked at missense mutations in SARS-CoV-2 genomes circulating in the United States, they found most mutations reached only the 5% threshold at the state level — with the exception of mutations associated with the B.1.1.7, B.1.427, and B.1.429 lineages.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.