The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is mediated by the fusion of the spike protein to nearby healthy cells. New research by German researchers sheds light on how this happens.
Sequential binding of two short amphipathic helices allows the SARS-CoV-2 fusion peptide to rapidly insert itself into the target membrane. They also found membrane fusion required strong anchoring made possible through double attachment and a conserved disulfide bridge.
The findings could help design fusion inhibitors to prevent viral entry and future biotechnological development of membrane anchors and fusogens for drug delivery applications.
The study “Binding of SARS-CoV-2 fusion peptide to host membranes” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
Different binding modes of the SARS-CoV-2 fusion peptide
The researchers performed several molecular simulations to evaluate several binding modes the SARS-CoV-2 fusion peptide can undertake during infection. The first set of simulations looked at the spontaneous binding of the fusion peptide to cell membranes. They positioned the fusion peptide nearby membranes that mimicked the endosome and outer leaflet of the plasma membrane.
The second set of simulations evaluated how strong the fusion peptide anchoring was during fusion. The researchers forced the fusion peptide away from an already bound membrane until it disconnected.
Formation of two short amphipathic helices
The SARS-CoV-2 fusion peptide after S1 shedding contains one highly conserved N-terminal amphipathic helix, a less conserved second amphipathic helix, and a C-terminal helix.
When the fusion peptide was exposed to water, the C-terminal residues, N824 and K825, of the N-terminal amphipathic helix unfolded and resulted in a shorter but more stable helix. The C-terminal end of fusion peptide also showed short disordered regions in three helical segments.
When exposed to an aqueous solution, the fusion peptide formed a single contiguous helix, the second segment unfolded, and the third segment retained its helix form.
The less conserved second amphipathic helix and the C-terminal helix conjoined together through a short loop. They also appeared connected through a conserved disulfide bridge.
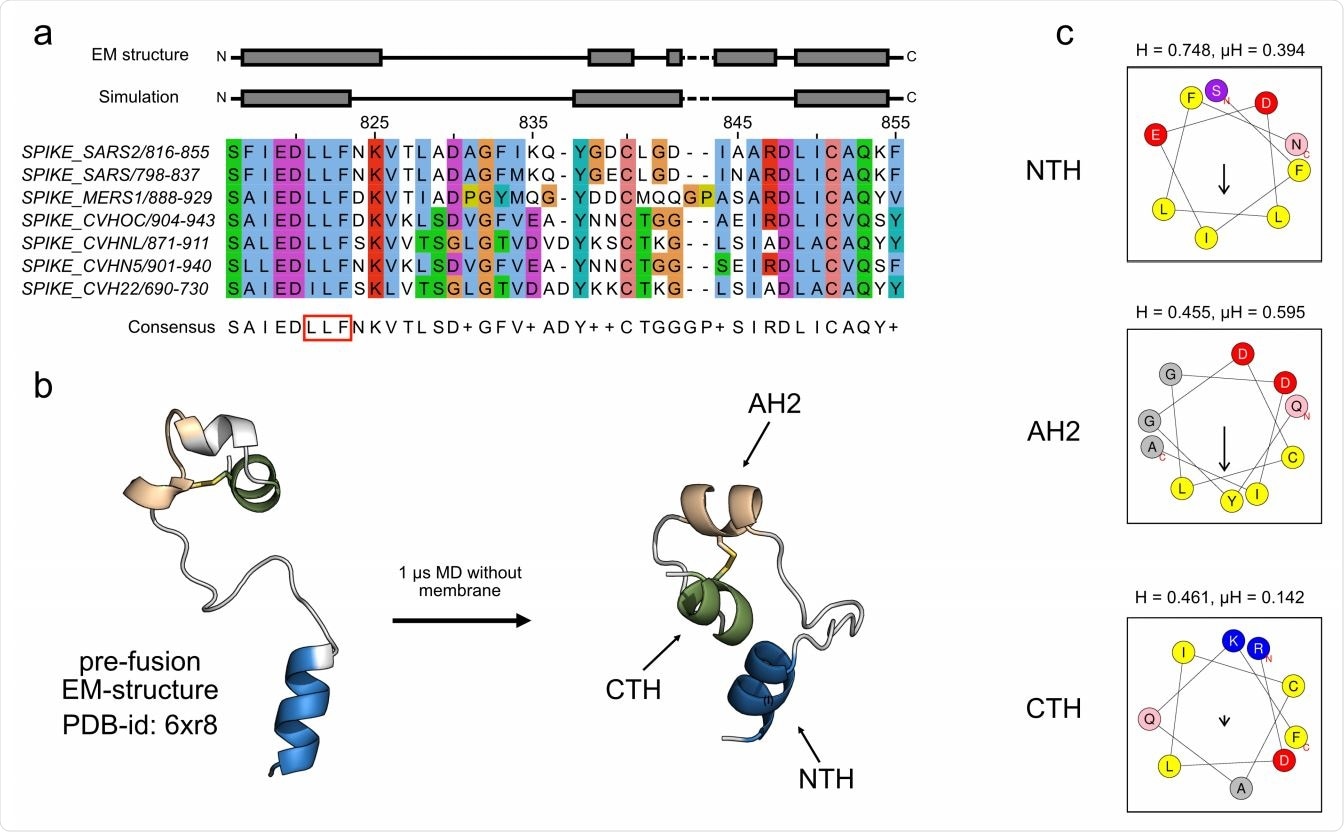
Amphipathic helices in SARS-CoV-2 fusion peptide. (a) Alignment of the FP region of human infectious coronaviruses and helix assignment from the cryo-EM structure (PDB ID: 6XR8, top)8 and from our simulation of the peptide without a membrane present (bottom). The red box marks the LLF motif in the N-terminal region. The fully conserved cysteines are at positions 840 and 851. The alignment was calculated using Clustal Omega. (b) Structural change of the FP in a 1 µs simulation in water and NaCl in cartoon representation (NTH in blue, AH2 in beige, CTH in green). (c) Amphipathic profiles of the NTH (top), the AH2 (middle) and the CTH (bottom) from SARS CoV-2 with hydrophobicity H and hydrophobic moment µH calculated with HeliQuest.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
N-terminal amphipathic helix is a stable contact for membrane fusion
Next, the researcher looked at whether the N-terminal amphipathic helix and the less conserved amphipathic helix are essential for anchoring the spike protein to the host membrane for viral entry.
Their simulations confirmed the N-terminal amphipathic helix inserted itself into the membrane. Four simulations showed insertion in the endosomal membrane, with three instances showing the N-terminal amphipathic helix was needed to maintain stability during fusion.
The researchers observed the N-terminal amphipathic helix was firmly bound to the membrane and remained bound for the entire simulation.
Two other insertions naturally occurred when the less conserved amphipathic helix and the C-terminal helix contacted the host membrane.
When the N-terminal amphipathic helix was the first binding towards the host membrane, F817 would then infiltrate below the phosphate headgroup region of the membrane.
The stability of the N-terminal amphipathic helix occurred because F823 would flip its orientation to the endosomal membrane. This allowed for a deeper insertion of the N-terminal amphipathic helix because three of its residues would be buried deeply in the membrane.
Membrane binding of the C-terminus end of the fusion peptide
The binding of the C-terminal involved the less conserved second amphipathic helix and the C-terminal helix with flexible elements at both ends of the amphipathic helix.
Creating stable contact with the host membrane involved short hydrophobic stretches inserted around residues I834, L841, and I844. Their position at the borders of the helices allowed for the additional insertion of residues.
High mechanical strength of N-terminal amphipathic helix binding
Removing the N-terminal amphipathic helix from the host membrane required great mechanical force. Unlike the C-terminal end, the N-terminal amphipathic helix resisted pulling forces of 40 to 65 pN.
A high force was needed to pull the N-terminal amphipathic helix when deeply bound with the inserted F823 residue. The estimated force to completely detach the helix from the deep state was 10 to 15 pN higher than when shallowly bounded.
Having all three helices in the host membrane greatly stabilized the fusion peptide membrane anchoring. Mechanical forces ranging from 130 to 254 pN were needed to detach all of them, indicating a fully bound fusion peptide is two to three times stronger than the binding of the N-terminal helix alone.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources