The pandemic of coronavirus disease 2019 (COVID-19), declared to be such in March 2020 by the World Health Organization (WHO), is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Seemingly unpredictable in the intensity of consequent illness, and with a terrifying capacity to spread both rapidly and extensively during asymptomatic infection – or in the presymptomatic phase of symptomatic disease – COVID-19 has attracted intensive research.
Symptomatology
The signs and symptoms of COVID-19 range from non-specific features like fever, muscle pain and tiredness, to organ- or system-specific symptoms like cough, diarrhea, headache, nausea, vomiting, and disturbances of taste and smell.
Computerized tomography of the chest showed bilateral ground-glass opacities in several lobes, with consolidation and thickening of the adjacent pleura.
People at high risk of severe illness following infection are more likely to be males, elderly, and with comorbidities such as diabetes, hypertension and heart disease.
Genomic features
The bat coronavirus RaTG13 has over 96% sequence identity with SARS-CoV-2. Intermediate hosts are likely to be involved, but none have been identified so far.
SARS-CoV-2 has typical genomic characteristics of a betacoronavirus, such as the Spike (S) gene, Nucleocapsid (N) gene, replicase complex (orf1ab) gene, Envelope (E) gene, and Membrane (M) gene, and two untranslated regions, the 5’ and 3′ UTRs.
The spike antigen is >93% identical in sequence to RaTG13 S, but shows seven differences from that of the previous highly pathogenic coronavirus, SARS-CoV, both in the N-terminal domain, and in the receptor-binding motif.
Again, the orf8 of the novel coronavirus may encode a structurally different protein from that of SARS-CoV-2. Deletions in this region were common during the SARS-CoV epidemic, and, in SARS-CoV-2, are associated with mild illness.
The mammalian zinc finger antiviral protein (ZAP) binds to the CpG site in the viral RNA and triggers its breakdown. The large CpG defect in SARS-CoV-2 may indicate that its intermediate and ancestral hosts had high ZAP levels.
SARS-CoV-2 also demonstrates faster protein biosynthesis, and increased efficiency of gene expression, with the effective number of codon (ENc) values of the SARS-CoV-2 structural proteins being lower by 5–20.
Pathogenicity explained
The earliest studies showed that SARS-CoV-2 gained entry to host cells via the angiotensin-converting enzyme II (ACE2) through its spike protein. However, it also readily infected VeroE6 cells that expressed high levels of transmembrane protease serine 2 (TMPRSS2), which primes the spike.
The inhibition of lung cell infection in the presence of TMPRSS2 inhibitor camostat mesylate suggests its potential use in COVID-19.
The high levels of TMPRSS2 in secretory cells, type I alveolar epithelium (AT1), and ciliated cells, containing SARS-CoV-2 RNA, suggest that these were the primary target tissues. This receptor increases with age, corresponding to the known increase in severity of infection in older people.
Cathepsin L, as well as host furin, also primes the spike protein – the latter acting at the unique furin-like cleavage site in the S1/S2 domain. Spike cleavage facilitates virus-cell fusion for viral entry, as well as cell-cell entry for the spread of infection.
Endocytosis is essential for actual viral entry into the host cell, using membrane channels such as phosphatidylinositol 3-phosphate 5-kinase (PIKfyve).
Thus, ACE2, TMPRSS2 and furin are all targets for potential drug development.
How does ACE2 increase COVID-19 risk?
In East Asian populations, high levels of ACE2 variants are found in tissues, which could mean population-dependent susceptibilities to the virus.
Again, cancer patients, smokers, and those with chronic obstructive pulmonary disease (COPD) all appear to be at higher risk of COVID-19. ACE2 is overexpressed in some solid tumors and in the lungs of smokers or COPD patients.
Mouse studies show that ACE2 increases in a subset of secretory cells following exposure to cigarette smoke. These cells multiply under such circumstances, leading to an absolute increase in SARS-CoV-2-susceptible cells.
Overexpression of ACE2 with hypomethylation is also observable in patients with systemic lupus erythematosus. Besides the increased risk of infection, the immunopathology may drive the abnormal expression of interferon-regulated genes, causing severe inflammation and progressive disease.
Obesity is also linked to increased ACE2 expression, while diabetes and related genetic traits are associated with a higher expression of ACE2 in the lung tissue. This might explain the higher vulnerability of these individuals to the infection.
How ACE2 is related to COVID-19 symptoms
The level of ACE2 in each tissue corresponds to the immune response in that tissue. In older people, but not in younger people, ACE2 expression is positively associated with immune signatures in the lung tissue.
In the lung, ACE2- and TMPRSS2-expressing cells contain viral particles and show evidence of injury. Nausea, vomiting and diarrhea were found to be typical of COVID-19, and intestinal cells show high ACE2 levels.
Low ACE2 levels may be sufficient for infection since low-ACE2 enterocyte precursors were just as readily infected as enterocytes. Viral transmission may occur through the gut, triggering systemic as well as gut-related progression.
Acute kidney injury has also been commonly reported in COVID-19, and here too, both receptors are co-expressed in multiple kidney cell types. Similar findings have been reported for the nervous system, pancreas, heart and olfactory epithelium.
In short, receptor ACE2 mediates the entry of SARS-CoV-2 into host cells, and its expression pattern and level also play important roles in COVID-19 susceptibility and symptoms.”
Aberrant immunologic findings in COVID-19
It is thought that severe COVID-19 is mediated by a weaker adaptive immune response, coupled with dysregulated systemic inflammation, leading to organ dysfunction and, in some cases, death.
In severe COVID-19, compared to mild and recovering patients, females had higher serum IgG concentrations than males, which may correspond to the better outcomes in this group.
Within three weeks of symptom onset, IgM and IgG antibodies to the viral spike were found in 94% and 100% of COVID-19 patients. Both antibody subtypes were predictive of SARS-CoV-2 infection.
Protective antibodies were elicited in children with COVID-19. Both total and IgG antibodies to the viral nucleocapsid and spike receptor-binding domain (RBD) and IgG-positive B cells were raised three weeks from symptom onset.
Neutralizing antibodies
Monoclonal antibodies and convalescent plasma have been used to treat critical COVID-19. Intriguingly, one single-domain antibody from a llama immunized with the earlier coronaviruses, SARS-CoV and MERS-CoV (Middle East Respiratory Syndrome coronavirus), cross-reacted with SARS-CoV-2 RBD.
In bivalent form, this antibody (VHH-72-Fc) neutralized a pseudovirus expressing the SARS-CoV-2 spike antigen. “VHH-72-Fc might act as a potential therapeutic candidate for COVID-19.”
Several monoclonal neutralizing antibodies have been isolated from specific memory B cells from recovered COVID-19 patients, which neutralize the SARS-CoV-2 spike-expressing pseudovirus by preventing ACE2-RBD binding.
Some of these may cause steric hindrance rather than blocking the binding epitopes of the virus or receptor. Some are cross-neutralizing, such as S309, especially when combined with other monoclonal neutralizing antibodies.
Another monoclonal antibody called 47D11 seems to inhibit SARS-CoV-2 infection by a different mechanism, since it does not prevent the spike-ACE2 binding.
These monoclonal antibodies may be an effective means of preventing and treating COVID-19.”
Dysregulated lymphocytes
Lymphopenia is an important finding in COVID-19, the decrease being proportional to the severity. Total, B, T and NK cells are all reduced.
Low B and CD8+ T cell counts and a low ratio of CD4+ to CD8+ T cells indicating inflammation predict a poor response to treatment. High CD8+ T cell counts may indicate higher cytotoxic antiviral activity.
Other predictive measures include serum interleukin-6 (IL-6) and IL-8, which are inversely related to falling lymphocyte counts in severe COVID-19; and a rising neutrophil to lymphocyte ratio (NLR) indicating a greater risk of death in hospitalized patients.
Lower B cell counts predict prolonged viral RNA shedding from the respiratory tract. High fluorescent lymphocytes (HFL) increase with progressive disease. These are associated with activated B cells or plasma cells, perhaps enhancing plasma cell and specific antibody production.
T cells from COVID-19 patients show signs of exhaustion, including programmed death-1 (PD-1) and T-cell immunoglobulin and mucin-domain containing-3 (Tim-3). This rises in CD8+ T cells during progressive disease.
CD4+ T cells show lower levels of tumor necrosis factor-α (TNF-α) and IFN-γ in severe disease, but CD8+ T cells have higher granzyme B and perforin levels.
The molecular marker called CD94/NK group 2 member A (NKG2A) is overexpressed on cytotoxic T lymphocytes and NK cells in COVID-19 but decreases with effective treatment. This is accompanied by higher NK cells and CD8+ T cells. That is, “targeting NKG2A might prevent functional exhaustion of lymphocytes and thus contribute to COVID-19 recovery.”
COVID-19 patients also showed markedly higher cytokine release. IFN-stimulated genes were upregulated, especially chemokines such as CXCL17 and CXCL8, along with inflammatory markers such as C-reactive protein (CRP), ferritin, and D-dimer.
These cytokines and markers of inflammation predict severe disease and death. “Therefore, strategies to suppress inflammatory responses are particularly important in patients with COVID-19, especially in severe cases.”
Animal models
The SARS-CoV-2 infection has been described in various animal models such as macaque, ferret, mouse, and hamster – within many of which infection is mild. Macaques develop mild infection, with some showing diffuse alveolar damage (DAD).
Rhesus macaques developed effective immunity to the virus after infection or vaccination. The human monoclonal antibody CB6, and the nucleoside analog remdesivir, protected rhesus macaques against lung injury.
Ferrets were more symptomatic after SARS-CoV-2 infection, showing acute bronchiolitis and inflammation. New lipoprotein fusion inhibitors are given intranasally prevented viral transmission in these animals by preventing virus-cell fusion. So did the ribonucleoside analog inhibitor MK-4482/EIDD-2801 and the TLR2/6 agonist INNA-051. These may be potentially useful in COVID-19.
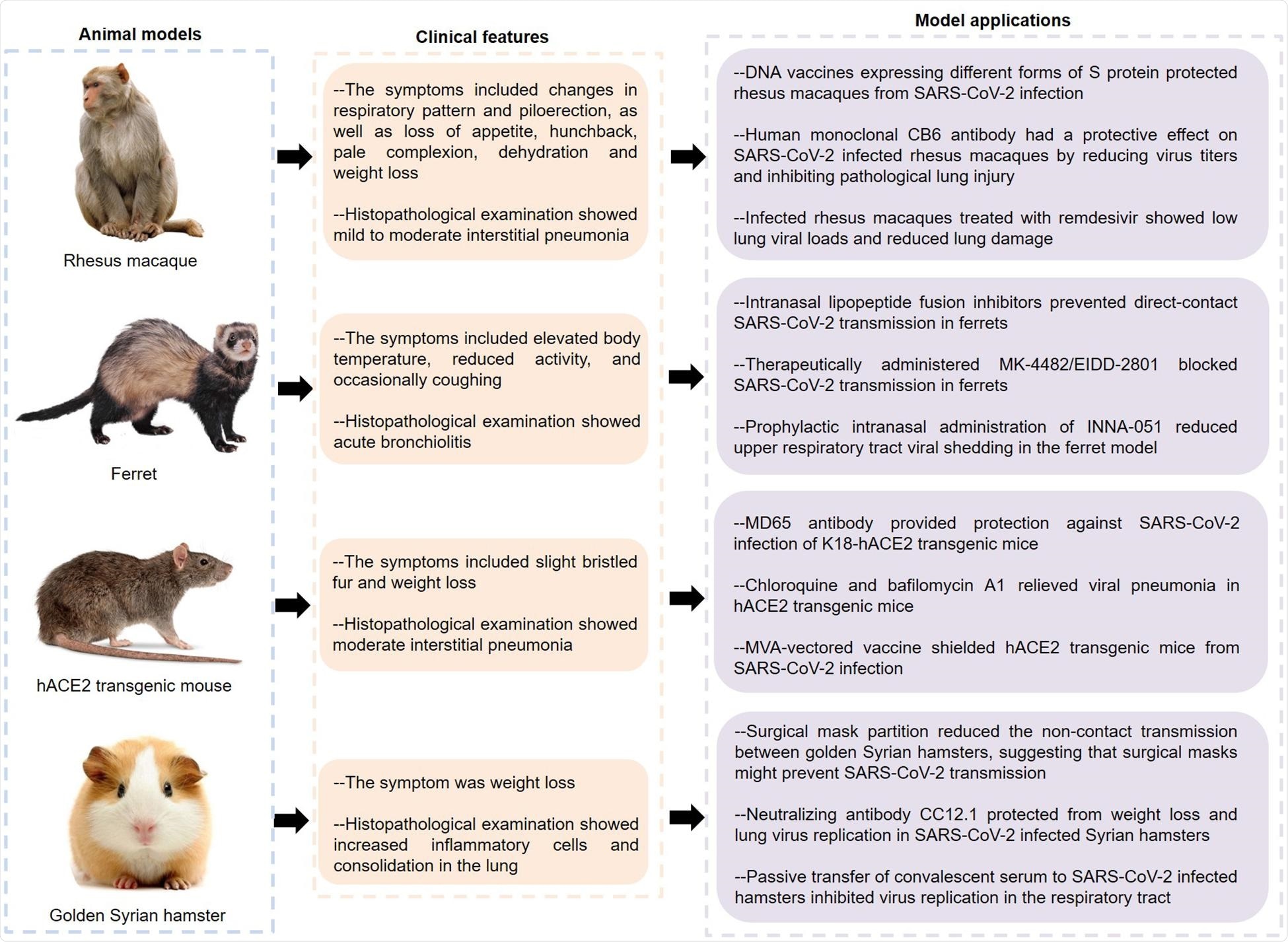
Features and applications of animal models of COVID-19. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; hACE2, human angiotensin converting enzyme II; COVID-19, coronavirus disease 2019.
Conversely, better-known drugs such as lopinavir-ritonavir, emtricitabine-tenofovir, and hydroxychloroquine sulfate were not found to be useful in treating infected ferrets, indicating the need for further study.
Mice showed interstitial pneumonia following spike-ACE2 interaction. Golden Syrian hamsters also showed symptoms of severe inflammation, and had olfactory involvement. These animals showed the same type of respiratory spread as in humans, making them good models for the infection.
The human monoclonal MD65 antibody, and certain endosomal acidification inhibitors, such as chloroquine, NH4CL, and bafilomycin A1, also showed antiviral effects in infected mice. The hACE2 mouse model is both cost-effective and practical and has thus been extensively used in COVID-19 research.
The anti-RBD antibody CC12.1 proved protective in hamsters, as did convalescent serum. Some felines and canines are also susceptible to the disease and may help model the infection in the future.
Future directions
Much remains to be known, including the efficacy of novel approaches to neutralizing the virus, indicating the need to understand how the infection occurs at the molecular level. While vaccines are being rolled out in many countries, the pace is often slow. Also, new variants threaten control by vaccination by resisting neutralizing antibodies elicited by vaccines.
In this context, it is of great significance to develop broadly neutralizing antibodies and vaccine boosters that target these SARS-CoV-2 variants.”