Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the causative agent of the coronavirus disease 2019 (COVID-19) pandemic, is a relatively recently identified coronavirus that has since spread around the globe.
SARS-CoV-2 is believed to be zoonotic in nature, in that the virus likely spilled over from an animal to a human host at some point in late 2019. Although the exact origin of this virus remains unknown, it is highly likely to have originated from horseshoe bat or pangolin coronaviruses. Both of these zoonotic strains of coronaviruses bear a large number of similarities with SARS-CoV-2 in different regards; therefore, ratifying which of these initially gave rise to SARS-CoV-2 remains a challenge.
In a recent study (not peer-reviewed*) published on the bioRxiv* server, Peng Zhou, Zheng-Li Shi and colleagues at the Wuhan Institute of Virology, with collaboration from scientists at the University of Chinese Academy of Sciences in China, have identified a new lineage of SARS-related coronaviruses that utilize bat angiotensin-converting enzyme 2 (ACE2) receptors for cell entry, which is the same receptor used by SARS-CoV-2 to infect host cells.
RaTG13, RatG15, and other SARS-related coronaviruses
ACE2 is an enzyme located on the cell membrane of primarily respiratory, cardiac, and liver cells in animals and is responsible for controlling blood pressure. SARS-CoV-2 uses ACE2 as an entry site to host cells. Since ACE2 is found most predominantly in the lungs and respiratory system, SARS-CoV-2 and, subsequently, COVID-19 often cause respiratory symptoms to arise following the infection.
Guo and colleagues sampled bats caught from the Yunnan province in 2015 and took anal swabs to extract viral ribonucleic acid (RNA), which were then stored and sequenced. Of the viruses that were found to exist within these bats, the team identified a viral strain, RaTG13, as being highly similar to SARS-CoV-2, sharing a 96.2% genomic identity.
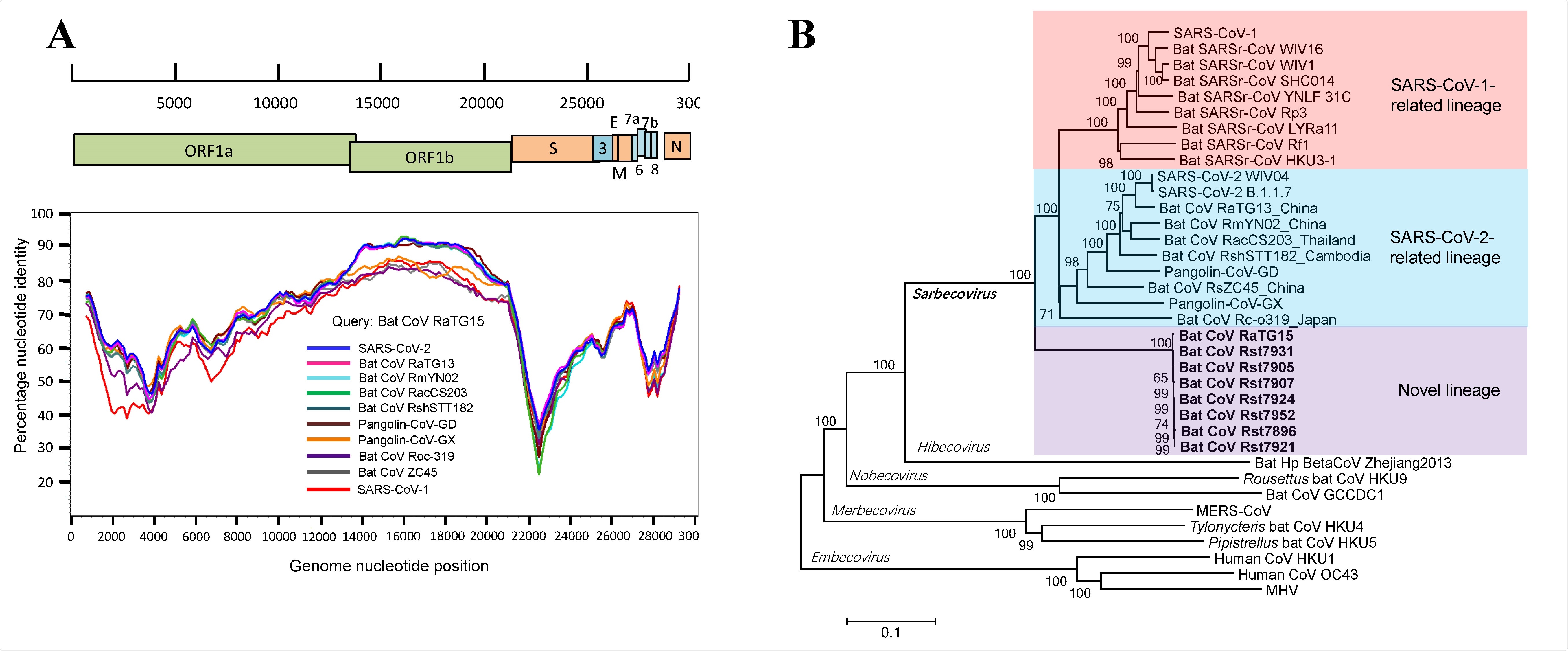
Discovery of a novel lineage of bat SARSr-CoVs. (A) Similarity plot analysis based on the full-length genome sequence of bat SARSr-CoV RaTG15. Full409 length genome sequences of SARS-CoV-1, SARS-CoV-2, bat and pangolin CoVs related to SARS-CoV-2 were used as reference sequences. The analysis was performed with the Kimura model, a window size of 1500 base pairs and a step size of 150 base pairs. (B) Phylogenetic tree based on complete genome sequences of betacoronaviruses. The trees were constructed by the Neighbour-joining method using the Jukes-Cantor model with bootstrap values determined by 1000 replicates. Bootstraps > 50% are shown. The scale bars represent 0.1 substitutions per nucleotide position. The novel SARSr-CoVs characterized in this study are shown in bold. Ra, Rhinolophus affinis; Rst, Rhinolophus stheno; Rsh, Rhinolophus shameli; Rs, Rhinolophus sinicus; Rac, Rhinolophus acuminatus; Rm, Rhinolophus malayanus; Rc, Rhinolophus cornutus; MHV, murine hepatitis virus.
Eight additional SARS-related coronaviruses were also found to share 93.5% of their genome identity to SARS-CoV-2. These eight viruses were all almost identical, as they shared more than 99.7% of their genome identity among each other.
One of these strains, RaTG15, was found to be 95.3% similar to SARS-CoV-2, and 92.5% similar to SARS-CoV-1, which was the causative agent of the SARS outbreak in 2002 to 2003. Analysis of the spike protein sequence of RaTG15 and the other strains showed deviation from other SARS-related coronaviruses, lacking deletions present in SARS-CoV-2 that determine ACE2 usage.
Through further tests, the research team concluded that RaTG15 and its related strains are unable to have the zoonotic potential of SARS-CoV-2. This contrasts with pangolin coronaviruses, many of which have a very high spillover potential in terms of their cell receptor usage.
Conclusion
This new lineage of SARS-related bat coronaviruses sheds light on the potential origins of SARS-CoV-2. RaTG13 and the other SARS-related coronaviruses discussed in this article demonstrated a minimal binding affinity for human ACE2, indicating that these viruses are unlikely to have given rise to SARS-CoV-2. Cross-species potential for infection is significantly higher in pangolin-related coronaviruses, despite the high (~99%) degree of relatedness between SARS-CoV-2 and this novel bat-CoV lineage.
The researchers of this study highlight that more longitudinal sampling of bats and pangolins, as well as other intermediate animal hosts, is required to fully grasp the origins of the COVID-19 pandemic.
Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.