The continued transmission of SARS-CoV-2 has led to its mutating several times. Many of the new mutations are on the virus spike protein, which has helped viral fitness and survivability by making it more infectious and giving it the ability to evade the immune system and neutralizing antibodies.
Several variants of concern have emerged in different parts of the globe, like the B.1.1.7 in the United Kingdom, the B.1.351 in South Africa, and the P.1 strain in Brazil. Two new variants have recently been reported from India, the B.1.617 and B.1.618 strains, characterized by several mutations in the spike protein.
The frequency of the B.1.617 variant in the population increased significantly beginning February 2021. This variant was reported to be seen in the United Kingdom, United States, and Singapore in later February 2021, spreading to more than 60 countries by May 2021.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In a study published on the medRxiv* preprint server, researchers report the emergence of a new strain of the SARS-CoV-2 virus in the state of West Bengal in India.
Analyzing genome sequences
The researchers obtained full genome sequences of 2,000 SARS-CoV-2 strains collected between January and March 2021 in India from the GISAID database. They performed a mutational analysis, obtained the amino acid sequence, and compared the sequences with the Wuhan strain.
After further analysis, the team found 70 strains with a new set of 11 mutations. There were three mutations in the non-structural proteins, three spike protein mutations, and two nucleocapsid protein mutations.
Sixteen of the sequences had the E484K mutation along with D614G, P681H, and V1230L mutations. These mutations fall in the GR clade, which has four other mutations that were also present in these strains. Further analysis of these variants revealed that these strains form a new cluster within the GR clade.
All the 70 sequences of this new strain have been seen only in the state of West Bengal. Apart from the 12 coexisting mutations, the researchers found 113 other different mutations.
Mutations may increase infectiousness
P681 is present next to the furin cleavage site, where proteolytic cleavage occurs between arginine (R685) and serine (S686). This site is at the junction between the S1 subunit, responsible for binding to the angiotensin-converting enzyme 2 (ACE2) receptor, and the S2 subunit, responsible for fusion between the cell membrane and the virus. Hence, the presence of a mutation at this site could affect S1/S2 cleavage and the infectiousness of the virus.
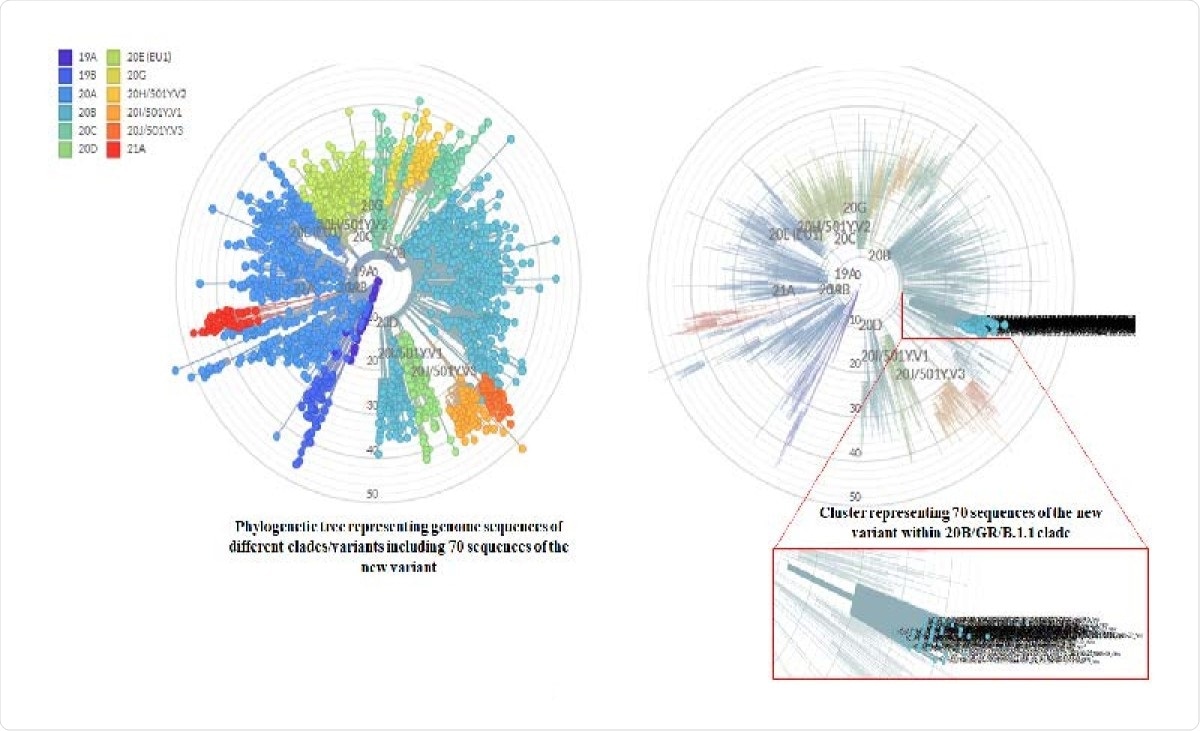
Molecular phylogenetic analysis of new variant strains by Nextcladebeta v.0.14.3. Left phylogenetic tree representing all the reference strains of different Nextstrain clades (Automatically selected by Nextclade) as well as 70 strains of the new variant. Each color depicting a different clade. Right phylogenetic tree highlighting only the 70 sequences of the new variant.
The P681H mutation has been seen before in the variant from the United Kingdom and is believed to increase spike protein cleavage, but does not seem to affect viral entry or its spread. The V1230L mutation has not been seen before and is located in the transmembrane region of the S2 subunit, which anchors the spike protein to the virus envelope. The substitution of valine with leucine could increase the binding of the spike protein to the viral envelope.
The new mutations could increase infectivity and transmissibility of the protein and may decrease the ability of antibodies to neutralize the virus. With higher infectivity rates reported in the second wave in India, emerging variants need to be monitored closely.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Sarkar, R. et al. (2021) Emergence of a new SARS-CoV-2 variant from GR clade with a novel S glycoprotein mutation V1230L in West Bengal, India. medRxiv. https://doi.org/10.1101/2021.05.24.21257705, https://www.medrxiv.org/content/10.1101/2021.05.24.21257705v1
- Peer reviewed and published scientific report.
Sarkar, Rakesh, Ritubrita Saha, Pratik Mallick, Ranjana Sharma, Amandeep Kaur, Shanta Dutta, and Mamta Chawla-Sarkar. 2022. “Emergence of a Novel SARS-CoV-2 Pango Lineage B.1.1.526 in West Bengal, India.” Journal of Infection and Public Health 15 (1): 42–50. https://doi.org/10.1016/j.jiph.2021.11.020. https://www.sciencedirect.com/science/article/pii/S1876034121004007.