A team of scientists from the University of Manchester, UK, has recently screened a panel of FDA-approved compounds to identify potential antiviral agents against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19). They have identified a group of kite-shaped compounds that are highly specific to SARS-CoV-1 and SARS-CoV-2. These compounds demonstrate high potency in inhibiting SARS-CoV-2 infection at early entry steps. The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
As of June 2, 2021, globally, there have been more than 171 million confirmed SARS-CoV-2 infections, with over 3.56 million deaths. The newly emerging viral variants have led to global concerns about the long-term efficacy of the COVID-19 vaccines that have been developed against the original strain of SARS-CoV-2. In order to effectively control infection caused by novel viral variants, novel therapeutics and prophylactics are necessary.
In the current study, the scientists have aimed to identify potential antiviral compounds that can specifically target SARS-CoV-2 at the entry steps. To screen a panel of FDA-approved viral entry inhibitors and natural compounds, they have utilized a pseudovirus system wherein mouse leukemia virus (MLV) is pseudotyped with the spike protein of SARS-CoV-2.
Screening of compounds
By infecting multiple cell types with pseudoviruses, the scientists identified human kidney epithelial cells stably expressing human angiotensin-converting enzyme 2 (ACE2) as the most suitable cell type for compound screening.
By screening 1,363 FDA-approved compounds and 137 natural compounds, they identified a specific group of structurally related entry inhibitors. The shape of these compounds was identical to the traditional Chinese kite. The compounds exhibited a highly conserved tri-cyclic core structure and an extension from the central 6- or 7-membered ring.
The final screening led to the identification of 61 kite-shaped compounds; of which five were excluded because of significant cytotoxic effects.
SARS-CoV-2 pseudotyped virus in anti-viral drug screening. Pseudovirus was generated on a mouse leukemia virus (MLV) backbone using a three plasmid system consisting of an expression vector for MLV gag and pol, a transfer vector carrying a luciferase reporter gene and an expression vector encoding the SARS-CoV-2 spike protein. Pseudovirus was used to infect 293T cells stably expressing the human angiotensin-converting enzyme 2 (ACE2). Pseudovirus entry was mediated by the binding of the spike protein to the ACE2 which then undergoes receptor-mediated endocytosis to trigger endosomal fusion to release the luciferase reporter gene into the cell cytoplasm. Infectivity was measured as luciferase read-out. Drugs from two APExBIO libraries were screened for their ability to inhibit infectivity by measuring the reduction in luciferase activity. Since the spike protein only mediates virus entry, the pseudovirus system could be used to identify drug hits that inhibit SARS-CoV-2 entry steps only. Drug cytotoxicity was measured using an XTT viability assay in non-infected cells. Drug images were obtained from PubChem and APExBIO.
Inhibition of SARS-CoV-2 infection
Based on the inhibitory potential, the scientists selected nine kite-shaped compounds for further evaluation. By using MLV-based pseudovirus systems of SARS-CoV spike, SARS-CoV-2 spike, and Middle East respiratory syndrome coronavirus (MERS-CoV) spike, they observed that all selected compounds had comparable efficacy in preventing infection by tested coronaviruses. Specifically, the selected compounds showed 1.2 to 8-fold higher efficacy in inhibiting SARS-CoV and SARS-CoV-2 than that of MERS-CoV, indicating that the inhibitory potency of these compounds depends on the types of host cell receptor that mediate viral entry.
Using different concentrations (10 mM to 0.05 mM) of selected compounds, the scientists developed dose-response curves to determine inhibitory efficacy. The findings revealed that at the concentration range of 1.9 mM to 4.7 mM, the kite-shaped compounds were effective in inhibiting 50% of the viral infection process (IC50). Importantly, they observed that even at the highest concentration, these compounds showed an acceptable cytotoxic profile.
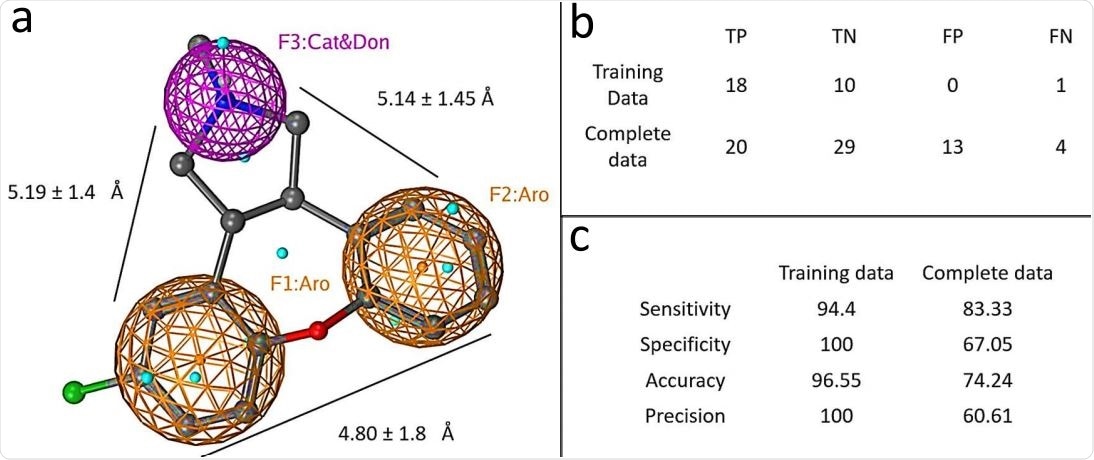
Graphical depiction of pharmacophore model. a) A three point pharmacophore model based on the kite-shaped molecules. The asenapine structure is superimposed (ball and stick representation) for comparison. Brown mesh represents aromatic moieties (Aro) and magenta mesh represents a H-bond donor/cation group (Cat&Don). Small spheres (cyan) highlight features in asenapine that are not relevant for the overall pharmacophore. (b) Displays the numbers of true (T) and false (F) positive (P) and negative (N) hits within the datasets that are discriminated by the pharmacophore (see Methods). Panel (c) summarises the pharmacophore model performance.
Mode of action of kite-shaped compounds
The scientists conducted time-of-addition experiments wherein the selected compounds were added to the cells at different time-points during SARS-CoV-2 infection. The findings revealed that the compounds effectively inhibited viral infection when added during the entry step. In contrast, the majority of the compounds failed to inhibit infection when added 1 and 2 hours after infection. These observations indicate that the kite-shaped compounds act as specific SARS-CoV-2 entry step inhibitors.
For further evaluation, they conducted temperature shift experiments to differentiate between viral attachment and post-attachment steps. The main principle of this experiment is that at a low temperature, the viral attachment step can proceed, but the post-attachment steps cannot proceed.
The findings of temperature shift experiments revealed that the kite-shaped molecules only moderately inhibited the attachment between viral envelop and host cell membrane. In contrast, the compounds showed the highest inhibitory potency when tested under conditions suitable for the post-attachment entry step.
Study significance
The study identifies a group of kite-shaped compounds that specifically target the post-attachment step of SARS-CoV-2 host cell entry, a highly conserved step in the viral lifecycle. Based on the antiviral mode of action, the scientists believe that these compounds can be used as broad-spectrum antiviral agents against newly emerging variants of SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources