Researchers in the UK and Austria have identified a novel class of antibodies that is highly effective at neutralizing the original strain of severe acute respiratory syndrome (SARS-CoV-2) and maintaining blocking activity against certain viral variants that have emerged.
The SARS-CoV-2 virus is the agent responsible for the ongoing coronavirus disease 19 (COVID-19) pandemic that has now claimed the lives of more than 3.76 million people.
The team identified ten single domain shark variable new antigen receptor (VNAR) antibody fragments that were highly effective at blocking the binding of the SARS-CoV-2 surface spike protein to its human host cell receptor angiotensin-converting enzyme 2 (ACE2).
The VNAR antibodies blocked the interaction between the receptor-binding domain (RBD) of the original Wuhan-Hu-1 spike protein and the ACE2 receptor. The antibodies also blocked this interaction when the spike proteins tested contained mutations found in the B.1.351 variant that emerged in South Africa and the P.1 variant that emerged in Brazil.
The team – from Ossianix Inc in Hertfordshire, Karl-Franzens-University in Graz and Medical University Graz – says that owing to their low complexity, small size, and formatting flexibility, single domain VNAR antibodies should be a useful adjunct to existing antibody approaches used in the treatment of COVID-19.
“VNAR antibodies directed against the SARS-CoV-2 spike protein expand the molecular toolbox of novel therapeutic approaches against COVID-19,” writes Pawel Stocki and colleagues.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The progress so far in combatting COVID-19
The initial step of the SARS-CoV-2 infection process is mediated by the viral spike protein when its RBD attaches to the host cell receptor ACE2.
The extracellular domain of the spike comprises two subunits. The S1 subunit contains the RBD (S1-RBD), which makes direct contact with ACE2, while the S2 subunit drives host cell entry by enabling viral fusion to the cell membrane.
The S1-RBD is the primary target of neutralizing antibodies following SARS-CoV-2 infection. This domain, therefore quickly became the main focus of efforts to develop vaccines and antibody therapeutics for the prevention and treatment of COVID-19.
A number of highly effective vaccines that have received emergency use authorization by national drug regulatory authorities are currently being rolled out in many countries across the globe.
The threats variants pose to future progress
However, a concerning development during the COVID-19 pandemic has been the emergence of SARS-CoV-2 variants harboring mutations that increase transmissibility or enable evasion of the host immune response.
For example, the N501Y mutation present in the B.1.1.7, B.1.351 and P.1 variants that emerged in the UK, South Africa and Brazil, respectively, has been shown to enhance binding affinity to ACE2 and to increase infectivity and virulence.
Similarly, the E484K mutation found in B.1.351 and P.1 enhances ACE2-binding affinity and has increasingly been associated with the reduced effectiveness of vaccines and therapeutic antibodies against these variants.
“The emergence of new variants is driven by mutation-prone viral replication and selective mechanisms involving the human host,” writes Stocki and colleagues.
Therefore, emerging novel variants will be naturally selected for their ability to infect and reinfect hosts and escape the immune response.
Consequently, vaccination and passive treatment with immunoglobulin- (Ig) based antibodies could add to this selection pressure and contribute to the emergence of new variants with the potential to evade host immunity.
How might VNAR antibodies help?
“An alternative therapeutic approach using single domain shark VNAR antibodies could be of immense value,” say the researchers.
These antibodies can bind to buried epitopes that are not available to conventional Ig-based antibodies. Furthermore, since they are not components of the human immune response, they are more likely to retain neutralizing activity against new variants that emerge under selection pressure.
“In addition, VNAR antibodies possess advantageous properties including small size, less complex structure, and a desirable chemical and thermal stability profile that allows for ease of manufacturing, storage without low-temperature refrigeration, and a variety of formulation options including the inhalation route,” writes the team.
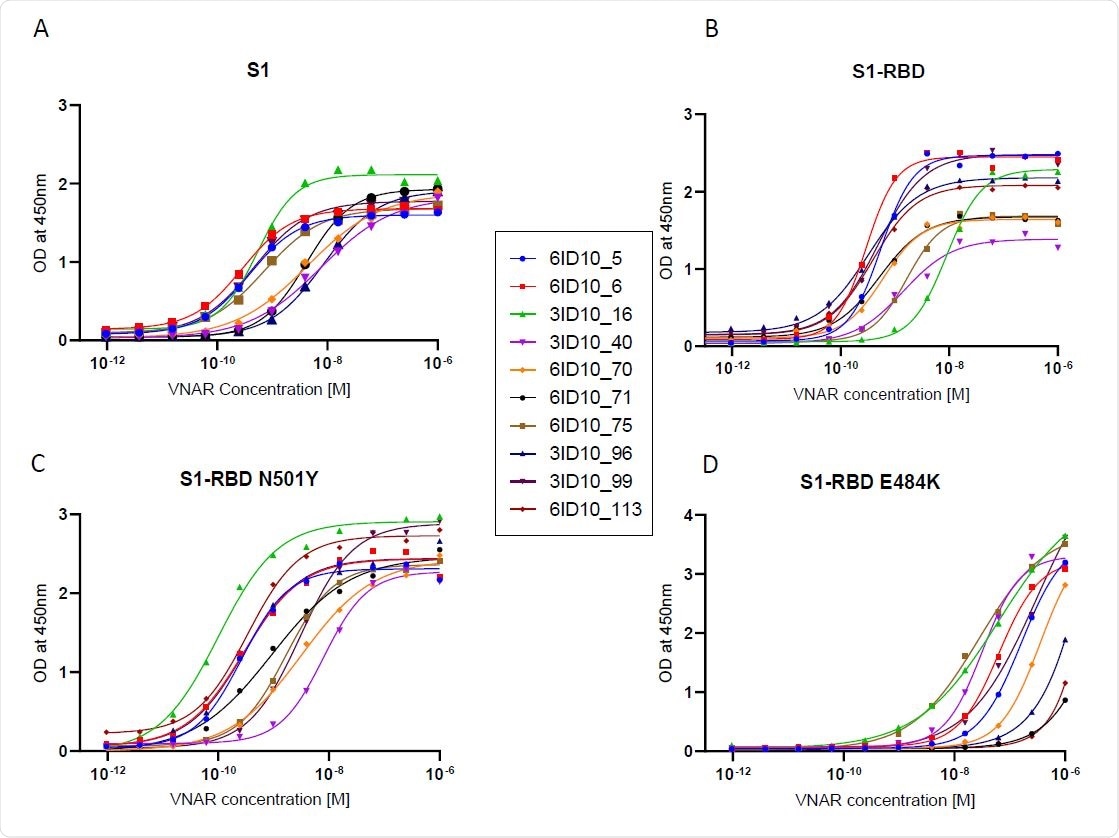
ELISA EC50 binding affinity of select VNAR-hFc antibodies to Wuhan and mutant spike proteins. ELISA was used to determine EC50 values for individual VNAR-hFc antibodies to (A) S1, (B) S1-RBD, (C) S1- RBD N501Y, and (D) S1-RBD E484K. Microplates were coated with different spike proteins and incubated with serially diluted VNAR-hFc antibodies followed by detection with HRP conjugated anti-human Fc antibody. The developed plates were read at 450nm and OD was used for 4-parametric non-linear regression model to calculate EC50 values.
What did the current study find?
The researchers targeted both the S1-RBD and the whole S1 subunit of spike protein from the Wuhan-Hu-1 variant to select neutralizing single domain VNAR antibodies using phage display.
These selection methods led to the identification of 56 target-specific binders that exhibited a high affinity for the spike protein.
The screens identified ten VNAR antibodies that were highly effective at blocking the interaction of recombinant spike protein with ACE2 in biochemical and cell-based assays and were further confirmed to block the activity of live Wuhan-Hu-1 virus in vitro.
The presence of the N501Y mutation in S1-RBD had no effect on the binding and blocking ability of the majority of VNAR antibodies tested.
In addition, three antibodies retained in vitro blocking activity when the E484K spike protein mutant was used.
“All 10 clones retained high-affinity binding and blocking activity against the S1- RBD N501Y mutant and 3 showed neutralizing activity against the S1-RBD E484K mutant,” write the researchers.
What did the authors conclude?
The team says this new and novel class of single-domain antibodies can be expected to retain neutralizing potential against viral variants carrying the N501Y or E484K substitutions.
“Single domain VNAR antibodies due to their low complexity, small size, unique epitope recognition and formatting flexibility should be a useful adjunct to existing antibody approaches to treat COVID-19,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Stocki P, et al. Single domain shark VNAR antibodies neutralize SARS-CoV-2 infection in vitro. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.06.08.447530, https://www.biorxiv.org/content/10.1101/2021.06.08.447530v1
- Peer reviewed and published scientific report.
Gauhar, Aziz, Cyril V. Privezentzev, Mykhaylo Demydchuk, Tanja Gerlza, Julia Rieger, Andreas J. Kungl, Frank S. Walsh, J. Lynn Rutkowski, and Pawel Stocki. 2021. “Single Domain Shark VNAR Antibodies Neutralize SARS‐CoV‐2 Infection in Vitro.” The FASEB Journal 35 (11). https://doi.org/10.1096/fj.202100986rr. https://faseb.onlinelibrary.wiley.com/doi/epdf/10.1096/fj.202100986RR.