Since the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported at the end of 2019, the virus has undergone numerous mutations, with the emergence of multiple variants. Some of these have shown increased infectivity, while others are resistant to the antibodies elicited by the parental strain or the vaccine. These are called variants of concern (VOCs).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
The variation between these strains and the parental strain is mainly due to the spike protein, which mediates both viral attachment to the host cell receptor – the angiotensin-converting enzyme 2 (ACE2) – and then undergoes proteolytic cleavage at the S1/S2 interface. This results in its being primed by the host enzyme TMPRSS2 and other proteases of the host cell to mediate virus-cell fusion as well as cell-cell fusion (syncytium formation).
This results in the entry of the virus into the cell to establish viral replication and productive infection. Spike mutations thus affect multiple processes, including binding affinity to ACE2, proteolytic cleavage, and incorporation of the spike into new viral particles, and the fit of antibody recognition sites.
The response of the host cell to the virus involves interferon (IFN) release, which is important in impeding viral entry and replication, as well as syncytium formation via interferon-induced transmembrane proteins (IFITMs) 1, 2 and 3. All these are broadly antiviral in their activity, probably enhancing membrane rigidity and preventing the crucial virus-cell fusion.
These proteins thus restrict infection by several enveloped viruses, including this one. This IFN response is thus a key part of innate immunity against viruses, limiting infection, and its impairment is therefore linked to more serious infection.
Replication comparable, fusogenicity increased
The researchers found that compared to the reference D614G strain, the alpha and beta strains showed equivalent rates of replication and virion release in various cell lines. All were equally inhibited by IFN-β1.
In a cell culture of human airway epithelial cells, the alpha/beta VOCs produced somewhat higher viral RNA, while the end results were similar in terms of infectious virion levels.
The alpha/beta strains did lead to the formation of measurably higher syncytia, at 4.5 and 3-fold, respectively, when identical doses of the virus were inoculated into the cells. Syncytia were equally inhibited for both variants when pre-treated with IFN-β1.
Interestingly, when the D614G, alpha- and beta-spike proteins were expressed in cells in culture, the latter two showed a doubling of syncytia formation, while the parental Wuhan strain was a little less fusogenic, relative to the D614G variant.
The findings indicate a greater degree of fusion of cells within the same period when exposed to the same dose of the alpha or beta variants relative to the D614G variant. The fastest syncytium formation occurred with the alpha variant, followed by the beta, relative to the D614G variant, while the introduction of the latter spike increased the fusogenicity of the Wuhan strain.
This reveals the key role played by the S1/S2 cleavage site in syncytium formation. However, no variant acquired resistance to IFITMs, which continued to reduce fusogenicity.
Different mutations, different effects
Among the individual mutations in each of these VOCs, the Δ69/70 deletion in the RBD of the alpha variant impaired syncytia generation, but this was opposed by the P681H and D1118H point mutations, especially the former which caused 2.5-fold higher fusogenicity compared to the D614G spike.
For the beta strain, the Δ242-244 deletion and the RBD point mutations K417N and E484K mutations led to reduced syncytium formation, while the N-terminal domain (NTD) mutation D251G had a small corrective effect. The E484K mutation reduced the fusogenicity of the alpha variant as well.
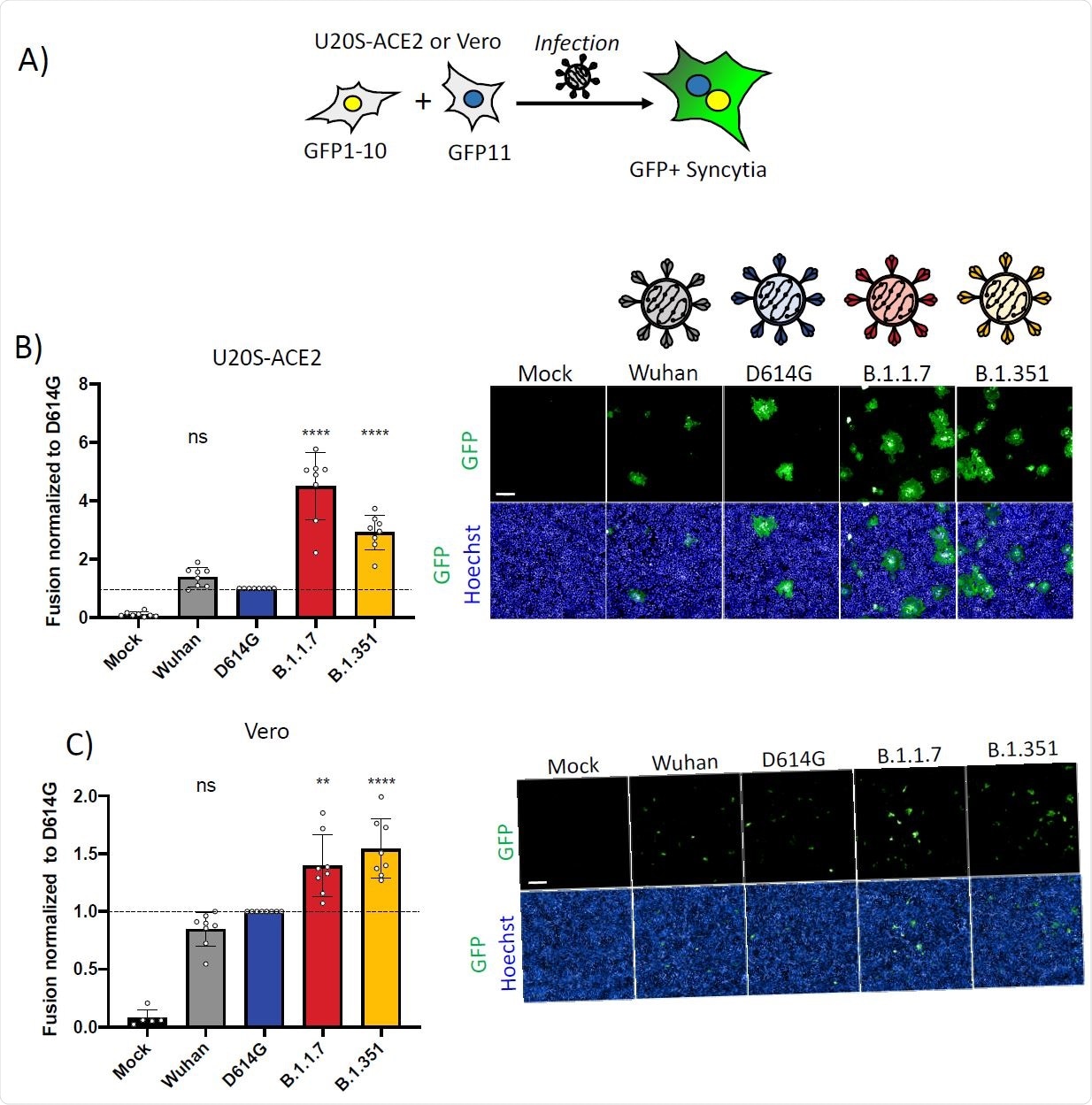
SARS-CoV-2 variant infection increases formation of syncytia in U2OS-ACE2 and Vero GFP-split cells. (A) U2OS-ACE2 or Vero cells expressing either GFP 1-10 or GFP 11 (1:1 ratio) were infected 24h after plating and imaged 20h (U2OS-ACE2) or 48h (Vero) post-infection. (B) Left Panel: Fusion was quantified by GFP area/ number of nuclei and normalized to D614G for U2OS-ACE2 20h post infection at MOI 0.001. Right Panel: Representative images of U2OS-ACE2 20h post infection, GFP-Split (Green) and Hoechst (Blue). Top and bottom are the same images with and without Hoechst channel. (C) Left Panel: Quantified fusion of Vero cells infected at MOI 0.01. Right Panel: Representative images of Vero cells 48h post infection, GFP-Split (Green) and Hoechst (Blue). Scale bars: 200 µm. Data are mean ± SD of 8 independent experiments. Statistical analysis: One-way ANOVA compared to D614G reference, ns: non-significant, *P < 0. 05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
What effect do these mutations have on ACE2 binding?
Binding affinity for the ACE2 receptor was highest with the alpha spike variant, with the beta variant second. The D614G and Wuhan variants had successively lower affinity.
The high affinity found with the alpha and beta variants is primarily related to the RBD N501Y mutation found in both. The K417N found only in the latter reduces ACE2 affinity. The E484K mutation has a very small positive effect on receptor binding, and the NTD Δ242-244 mutant a minor negative effect.
The spike variants also changed the fit of the epitopes or antibody binding sites. Thus, one monoclonal antibody (MAb) targeting the RBD of the spike protein failed to bind to the K417N mutant. A second RBD antibody showed a more diffuse failure of recognition, suggesting that the whole spike structure had changed.
Another MAb directed at the NTD did not recognize the alpha or beta VOCs, specifically failing to bind their NTD ΔY144 and Δ242-244 mutations.
What are the implications?
The researchers demonstrated that the alpha and beta variants show the same replication rates as D614G while being more fusogenic, producing more syncytia within the same period with the same viral dose. They were also more rapidly fusogenic, both these effects being related to their spike mutations.
However, innate immunity remained active against both variants, specifically IFN-β1, which inhibited infection by both. Moreover, the fusogenicity of both remained sensitive to IFITMs, contradicting recent reports that the alpha strain was more resistant to IFN-mediated innate immunity.
The high fusogenicity of the alpha variant is because of the many mutations with a fusogenic effect compared to the single deletion that reduces syncytium formation, in direct contrast to the mostly restrictive mutations in the beta variant. The researchers also identified P681H (like the P681R found in the B.1.617.2 and B.1.617.3 variants) to be the single most fusogenic mutation, acting at the S1/S2 interface to cleave the spike protein and promote fusion between cells.
The mutations that dramatically improved spike-receptor binding were not always associated with fusogenicity. For instance, with N501Y and D614G, increased cell-cell fusion was observed to increase only with the latter.
Finally, spike mutants that reduce fusogenicity, or at least leave it unchanged, successfully escape antibody recognition. The high-affinity N501Y mutant, on the other hand, leaves the virus susceptible to neutralizing antibodies. The RBD mutations E484K and K417N produce much lower increases in binding affinity for ACE2.
This indicates some degree of trade-off has occurred, which allows the virus to evade antibody-mediated neutralization even though its ultimate efficiency is impaired thereby. The successful spread of these strains may indicate the ability of the combination of mutations to evade recognition by neutralizing antibodies.
However, not all neutralizing antibodies restrict syncytial formation; some only inhibit virus-cell fusion. This may be because the former is more resistant to antibodies than the virus outside the cell.
The rise to dominance of the alpha B.1.1.7 strain may be due to the characteristics shown here, such as high affinity for the ACE2 receptor and increased fusogenicity, whereby it has overtaken the beta variant globally. Further study is required to understand if these are the sole reasons and the role played by mutations on these biological characteristics in newer variants now spreading rapidly worldwide.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Rajah, M. M. et al. (2021). B.1.1.7 and B.1.351 SARS-CoV-2 variants display enhanced Spike-mediated fusion. bioRxiv preprint. doi: https://doi.org/10.1101/2021.06.11.448011, https://www.biorxiv.org/content/10.1101/2021.06.11.448011v1.
- Peer reviewed and published scientific report.
Rajah, Maaran Michael, Mathieu Hubert, Elodie Bishop, Nell Saunders, Remy Robinot, Ludivine Grzelak, Delphine Planas, et al. 2021. “SARS‐CoV‐2 Alpha, Beta, and Delta Variants Display Enhanced Spike‐Mediated Syncytia Formation.” The EMBO Journal, October. https://doi.org/10.15252/embj.2021108944. https://www.embopress.org/doi/full/10.15252/embj.2021108944.