Researchers in the UK have warned that a widespread mutation that has arisen in the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) appears to escape recognition by CD8 T-cell responses in both convalescent patients and recipients of the current coronavirus disease 2019 (COVID-19) vaccines.
The spike mutation – P272L – has so far arisen in five different SARS-CoV-2 lineages, including the B.1.177 lineage that was associated with a second pandemic wave in Europe.
Now, a team from Cardiff University, Cardiff University School of Medicine, the University of Oxford, and the COVID-19 Genomics UK consortium have shown that CD8 T-cells from a cohort of convalescent patients that comprised more than 120 different T cell receptors (TCRs) failed to respond to the P272L variant.
Furthermore, sizeable populations of CD8 T-cells from individuals immunized with the currently approved COVID-19 vaccines failed to bind to a P272L reagent.
Andrew Sewell and colleagues say that viral escape at prevalent T-cell epitopes may be particularly problematic when vaccine immunity is focused on a single protein such as the SARS-CoV-2 spike.
The viral escape observed here provides a strong argument for the inclusion of multiple viral proteins in next-generation vaccines and highlights the urgent need for monitoring T-cell escape in new SARS-CoV-2 variants, says the team.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.
.jpg)
Study: Emergence of immune escape at dominant SARS-CoV-2 killer T-cell epitope. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The role of adaptive immunity in protecting against SARS-CoV-2
The host adaptive immune system protects against viral infection by selecting specific antigen receptors present on B-cells and T-cells.
While the mechanisms SARS-CoV-2 uses to escape adaptive immunity are not yet well understood, they are now firmly in the spotlight, says Sewell and colleagues.
Although most research into the immune responses generated by SARS-CoV-2 infection has so far focused on antibody-mediated immunity, some evidence suggests that antibodies may play a secondary role to T-cells in clearing the virus.
“There are also many reports of healthy individuals successfully controlling SARS-CoV-2 infection without having detectable neutralizing, or receptor-binding domain (RBD) antibodies, while having prominent SARS-CoV-2-specific T-cell memory,” write the researchers.
“Indeed, the relative scarcity of naïve T-cells in individuals over 65 years old and the connection between aging and impaired adaptive immune responses to SARS-CoV-2 has been suggested as a major cause of severe disease,” they add.
A growing body of evidence also indicates that Human Leukocyte Antigen class I (HLA-I)-restricted CD8 killer T-cells contribute to the immunity provided by currently approved COVID-19 vaccines.
What did the researchers do?
Given the potential importance of CD8 T-cells in adaptive immune protection against COVID-19, the team set out to examine dominant T-cell responses to SARS-CoV-2 infection via the most prevalent major histocompatibility complex (MHC) allele in humans – HLA A*02.
This HLA MHC class I molecule presents processed intracellular viral protein antigens at the cell surface for inspection by CD8 T-cells.
A previous study using overlapping peptides from the entire SARS-CoV-2 proteome identified that the most common HLA A*02-restricted responses among convalescent patients were to epitopes contained within residues 3881 to 3900 of the open reading frame 1ab (ORF1ab) and residues 261 to 280 of the spike protein. The spike epitope was then narrowed down to residues 269-277 (sequence YLQPRTFLL).
“We, therefore, focused our attention on residues 269-277 of the SARS-CoV-2 spike protein,” says Sewell and colleagues.
What did the study find?
The researchers found that the most prevalent mutation – P272L – in this T-cell epitope (sequence YLQLRTFLL) was not recognized by more than 120 TCRs that responded to the founder epitope (YLQPRTFLL) across a cohort of nine HLA A*02+ convalescent patients.
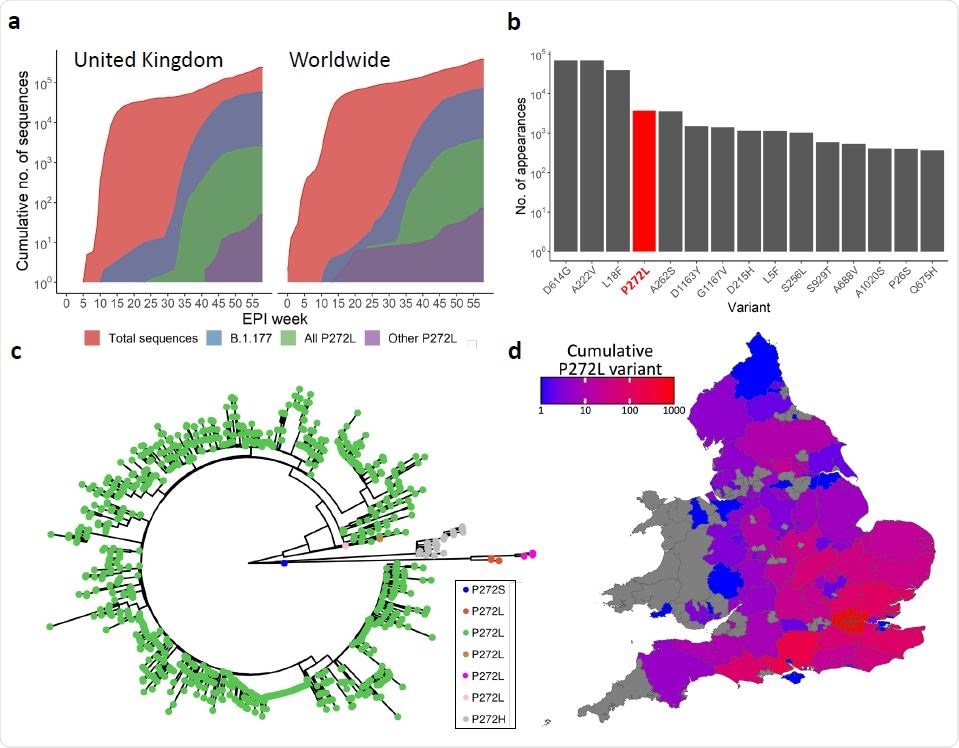
SARS-CoV-2 variation in the immunodominant Wuhan-YLQPRTFLL dominant HLA A*02:01- restricted CD8 T-cell epitope. a, Cumulative frequency of all sequences; sequences in the B.1.177 lineage; and sequences possessing the P272L variant, both in B.1.177 and other lineages across the United Kingdom (left) and worldwide (right). Total number of P272L variants by nation, binned into periods of 10 epidemiological (EPI) weeks is shown in Supplementary Figure 1. b, Top 15 most frequently observed variants observed in worldwide Spike glycoprotein sequence data in lineage B.1.177. Data for all lineages is shown in Supplementary Figure 3. c, Phylogenetic tree showing 1227 taxa with variants at Spike position 272, with colours indicating subtrees representing potential independent mutations, computed by ASR on a larger tree of 200 221 taxa (not shown). d, Total number of sequences possessing P272L variant in England and Wales per administrative region up to and including 31st January 2021.
The team also found that sizeable populations (0.01-0.02%) of CD8 T-cells that were stained with peptide-HLA A*02 multimers bearing the YLQPRTFLL peptide in a cohort of individuals vaccinated against SARS-CoV-2 could not be stained by reagents manufactured using the P272L variant.
“We conclude that the YLQPRTFLL-specific T-cells induced by current COVID vaccines fail to engage to the P272L variant sequence,” writes Sewell and colleagues.
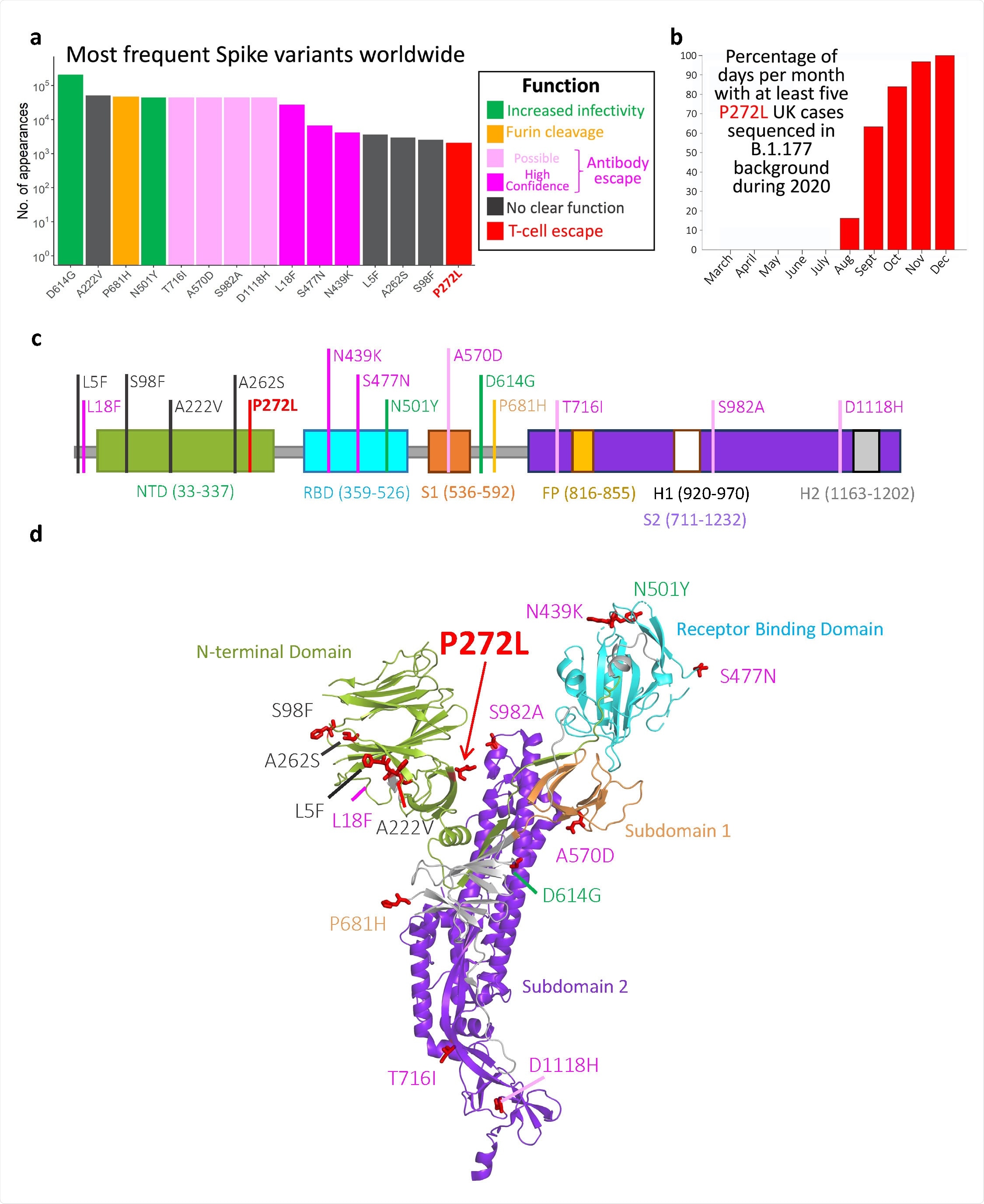
Top 15 most frequently observed variants observed in worldwide spike glycoprotein sequence data in all lineages. Those thought to be associated with enhanced viral transmission are shown in green and those listed as potentially escaping from antibody-mediated-mediated immunity are shown in light pink (possible) and dark pink (high confidence) ass assigned at http://sars2.cvr.gla.ac.uk/cog-uk/#shiny-tab-immunology. This color-coding of substitutions is used throughout the figure. Variants of unknown function in grey. N501Y also escapes from antibodies b, Fraction of days per month where five or more P272L variants were sequenced in B.1.177 background. c-d, Mapping of mutants shown in A onto Spike sequence map and prefusion structure (PDB 6VXX; Walls et al. 2020. Cell, 181, 281-291)
Important implication for the development of next-generation vaccines
The researchers say the findings show that SARS-CoV-2 can readily alter its spike protein via a single amino acid substitution so that it is no longer recognized by CD8 T-cells targeting the most prevalent spike epitope restricted by the most common HLA-I across the population.
“The specific focusing of immune protection on a single protein (e.g., SARS-CoV-2 spike favored by all currently approved vaccines) is likely to enhance any tendency for escape at predominant T-cell epitopes like YLQPRTFLL,” says the team.
“Our demonstration that mutations that evade immunodominant T-cell responses through population-frequent HLA can readily arise and disseminate strongly suggests that it will be prudent to monitor such occurrences and to increase the breadth of next-generation SARS-CoV-2 vaccines to incorporate other viral proteins,” concludes Sewell and colleagues.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Sewell A, et al. Emergence of immune escape at dominant SARS-CoV-2 killer T-cell epitope. medRxiv, 2021. medRxiv, 2021. doi: https://doi.org/10.1101/2021.06.21.21259010
- Peer reviewed and published scientific report.
Dolton, Garry, Cristina Rius, Md Samiul Hasan, Aaron Wall, Barbara Szomolay, Enas Behiry, Thomas Whalley, et al. 2022. “Emergence of Immune Escape at Dominant SARS-CoV-2 Killer T Cell Epitope.” Cell 185 (16): 2936-2951.e19. https://doi.org/10.1016/j.cell.2022.07.002. https://www.cell.com/cell/fulltext/S0092-8674(22)00849-2