The coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected more than 188 million people worldwide and has claimed more than 4 million lives as of July 15, 2021.
Several vaccines have already received emergency use authorization from the regulatory bodies in many countries around the world. In fact, current estimates indicate that over 3.53 billion vaccine doses have been administered worldwide, which is equal to 46 doses for every 100 people.
Despite the rapid rate at which vaccines are being administered, the emergence of SARS-CoV-2 variants has threatened the effectiveness of these vaccines. Therefore, robust therapeutic strategies remain extremely important to reduce the mortality rate of COVID-19.
 Study: Published anti-SARS-CoV-2 in vitro hits share common mechanisms of action that synergize with antivirals. Image Credit: Gorodenkoff / Shutterstock.com
Study: Published anti-SARS-CoV-2 in vitro hits share common mechanisms of action that synergize with antivirals. Image Credit: Gorodenkoff / Shutterstock.com
Background
Through an in vitro drug repurposing strategy that involves the utilization of previously approved drugs for COVID-19 treatment, scientists have identified 184 potential candidates. In fact, recent studies have found that several drugs approved by the United States Food and Drug Administration (FDA) can target various phases of the interactions that occur between SARS-CoV-2 and the host cell. To this end, the eIF4A inhibitor zotatifin and the eEF1A inhibitor plitidepsin have been found to damage the cap-dependent messenger ribonucleic acid (mRNA) translation of coronaviruses.
Most of the drugs that have been identified by this repurposing strategy have shown moderate antiviral activity. However, their mechanisms of action (MoAs) have not been clearly understood, owing to the haste to test these drugs in clinal trials. The failure of drugs such as chloroquine and ivermectin has highlighted major gaps in the research. More specifically, the development of effective therapeutics requires a more thorough understanding of the MoA of the drug, the virus’s life cycle, and virus-host interactions.
To this end, transcriptomic profiles provide a detailed picture of the drug-gene interactions that could be explored to further understand drug mechanisms. Assuming that drugs with similar MoAs possess comparable gene expression profiles, or specific drugs can restore the gene expression that gets dysregulated in the disease state, transcriptomic profiles could also be used to evaluate mirroring or reversal effect between drugs and diseases.
Analyzing transcriptomic profiles to develop new COVID-19 therapeutics
In a new study published in the journal Briefings in Bioinformatics, researchers collected 184 compounds that had been identified from 14 different studies to possess in vitro anti-SARS-CoV-2 activity. These compounds demonstrated inhibitory activity on both SARS-CoV-2 induced-cytopathic effect and SARS-CoV-2 replication in the host cells with EC50/IC50 values of less than 50 μM.
In this study, researchers observed that the compounds capable of regulating microtubule cytoskeleton organization pathways and cholesterol homeostasis can also be used to treat COVID-19. In other words, while screening for the SARS-CoV-2 drugs from the available databases, 36% of the positive hits showed common mechanistic effects via the regulation of cholesterol homeostasis and/or microtubule cytoskeleton organization.
The large-scale profiling of SARS-CoV-2 virus-host interactions revealed that viral proteins Orf8 and Orf9c can directly attach with NPC2 and SCAP, both of which are important components for the transportation and monitoring of cholesterol levels. Additionally, the ‘microtubule-based process’ was found to enhance the host protein targets of viral NSP10 and NSP13.
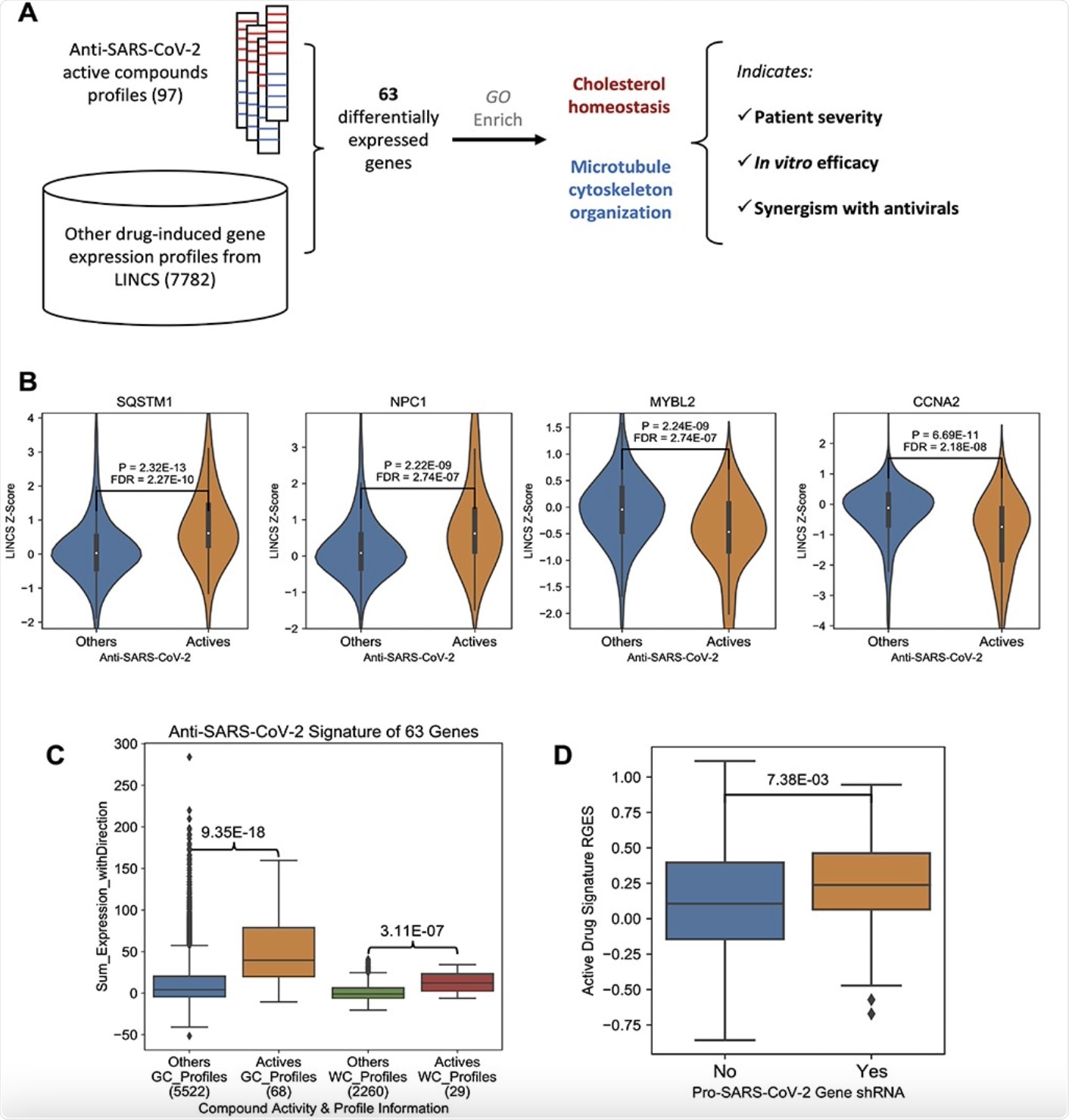 Anti-SARS-CoV-2 compound signature gene expression change. A, The workflow of this research. B, Example genes were induced (SQSTM1 and NPC1) or suppressed (MYBL2 and CCNA2) by anti-SARS-CoV-2 compounds. The y-axis indicates the LINCS z-score of a specific compound and a higher score means higher expression change. P-values were derived from Wilcox rank sum tests, and further corrected across all LINCS 978 genes. C, A boxplot showing the comparison of the effects on the 63 signature genes between active compounds and other compounds. The y-axis indicates an overall effect of a specific compound on the 63 genes, and a higher score means a better alignment with the up/downregulation pattern. On the x-axis, the number of gene expression profiles in each group is labeled under the group name. P-values were derived from Wilcox rank sum tests. D, A boxplot comparison between anti-SARS-CoV-2 CRISPR screening gene hits and non-hits. A higher RGES score on y-axis indicates a query host cell shRNA knockdown-induced gene expression profile more closely resembles the summarized gene expression signature of anti-SARS-CoV-2 active compounds. x-axis denotes whether a query host gene knock-out makes the cells resistant to SARS-CoV-2 infection. In the box plot, the central line represents the median value, and the bounds represent the 25th and 75th percentiles. The whiskers are 25th/75th quartiles plus 1.5 times the interquartile range.
Anti-SARS-CoV-2 compound signature gene expression change. A, The workflow of this research. B, Example genes were induced (SQSTM1 and NPC1) or suppressed (MYBL2 and CCNA2) by anti-SARS-CoV-2 compounds. The y-axis indicates the LINCS z-score of a specific compound and a higher score means higher expression change. P-values were derived from Wilcox rank sum tests, and further corrected across all LINCS 978 genes. C, A boxplot showing the comparison of the effects on the 63 signature genes between active compounds and other compounds. The y-axis indicates an overall effect of a specific compound on the 63 genes, and a higher score means a better alignment with the up/downregulation pattern. On the x-axis, the number of gene expression profiles in each group is labeled under the group name. P-values were derived from Wilcox rank sum tests. D, A boxplot comparison between anti-SARS-CoV-2 CRISPR screening gene hits and non-hits. A higher RGES score on y-axis indicates a query host cell shRNA knockdown-induced gene expression profile more closely resembles the summarized gene expression signature of anti-SARS-CoV-2 active compounds. x-axis denotes whether a query host gene knock-out makes the cells resistant to SARS-CoV-2 infection. In the box plot, the central line represents the median value, and the bounds represent the 25th and 75th percentiles. The whiskers are 25th/75th quartiles plus 1.5 times the interquartile range.
The utility of using ChoMCyto patterns
The authors designed a ChoMCyto score to quantify the reviewed drug effect pattern as it relates to the severity of COVID-19. When the ChoMCyto pattern was applied, the researchers could predict anti-SARS-CoV-2 efficacy. To this end, they determined that a new compound known as monensin inhibits SARS-CoV-2 replication with an EC50 of 11 μM in Vero-E6 cells.
Previous research has indicated that monensin inhibits viral transportation within the Golgi complex. This indirect antiviral mechanism inspired the authors of the present study to examine the synergism that exists between established antiviral drugs and repurposed drugs targeting host cellular ChoMCyto genes.
In this context, the researchers obtained a recent combination screening study from The National Center for Advancing Translational Sciences (NCATS) and found 15 host-targeting compounds were combined with some of the antivirals. Of these, the drugs with higher ChoMCyto scores were expected to present a synergistic effect with antivirals.
Conclusion
The present study concluded that cholesterol homeostasis and microtubule cytoskeleton organization pathways are disturbed by the COVID-19 infection. Among the potential anti-SARS-CoV-2 compounds that have been previously reported, one-third of them are inclined towards the restoration of biological processes inside the host cells.
The researchers emphasized that targeting both of these pathways might enhance the effectiveness of known antiviral drugs used for the treatment of COVID-19. These compounds, along with their cytotoxicity, efficacy, chemical structure, and MoAs have all been reported in this study.