Close contact between individuals has contributed enormously to the sporadic spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) across the world.
According to the United States Centers for Disease Control and Prevention (CDC), ‘close contact’ is defined as an individual who spent at least 15 minutes, over a 24-hour period, within 2 meters of an individual with coronavirus disease 2019 (COVID-19). This definition of close contact has become widely used in investigative studies on ‘contact’ and determining the average risk of COVID-19 in healthcare settings and communities.
 Study: Variation in SARS-CoV-2 bioaerosol production in exhaled breath. Image Credit: eldar nurkovic / Shutterstock.com
Study: Variation in SARS-CoV-2 bioaerosol production in exhaled breath. Image Credit: eldar nurkovic / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Researchers from the Stanford University School of Medicine, however, felt that the empirical basis for this specific time window is limited. In fact, the researchers hypothesized that individual characteristics and actions such as talking may also affect the overall bioaerosol shedding and impact exposure risk over this time window. To further study this hypothesis, the researchers quantified SARS-CoV-2 from exhaled breath samples to determine the impact of speech and individual characteristics on viral shedding.
Improving the sensitivity of mask sampling
The production of SARS-CoV-2 bioaerosols by an infected individual varies between individuals. As a result, bioaerosol production is generally calculated by modeling the combined estimates of concentrations in airway swabs with data on total droplet production volume.
This approach has largely been used due to the lack of a flexible bioaerosol sampling tool that can directly quantify the variation in SARS-CoV-2 bioaerosol abundance. While mask sampling has been explored as a method of bioaerosol sampling, preliminary studies have found the approach to produce marginal sensitivities.
In an effort to improve the sensitivity and sample recovery of mask sampling, a group of Stanford researchers recently developed a novel mask sampling tool to assess bioaerosol variation within individuals. After quantifying and sequencing \SARS-CoV-2 from exhaled breath samples, the researchers used this data to investigate the impact of speech and individual characteristics on viral shedding.
About the study
For this investigation, the researchers developed a mask sampling tool that included fitted N95 masks containing a 47 millimeter (mm) Petri dish with a gelatine membrane filter. A total of 141 mask samples were collected from 97 COVID-19 positive patients from Stanford Hospital between September 2020 and March 2021.
The patients were divided into two groups. Whereas Group A included patients who wore a mask for 30 minutes and were told to either talk or remain silent, Group B consisted of patients who were instructed to wear two masks for 15 minutes each. While wearing the first mask, Group B participants did not talk at all; however, while wearing the second mask, they talked with the interviewer or a family member.
Study findings
“Over 15 minutes of collection, total viral copies captured by masks varied substantially, ranging from 0 to 5.0 x 106 copies while talking to 0 to 8.4 x 104 copies while not talking.”
A moderate correlation was reported in individuals of a greater age who were in the 15-minute talking group and higher viral loads in exhaled breath samples. However, other factors including the duration of symptoms, the days from the first SARS-CoV0-2 test, and the number of human RNAseP copies were not found to affect the viral copies collected in the masks. Importantly, inpatients had significantly lower mask positivity rates as compared to outpatients.
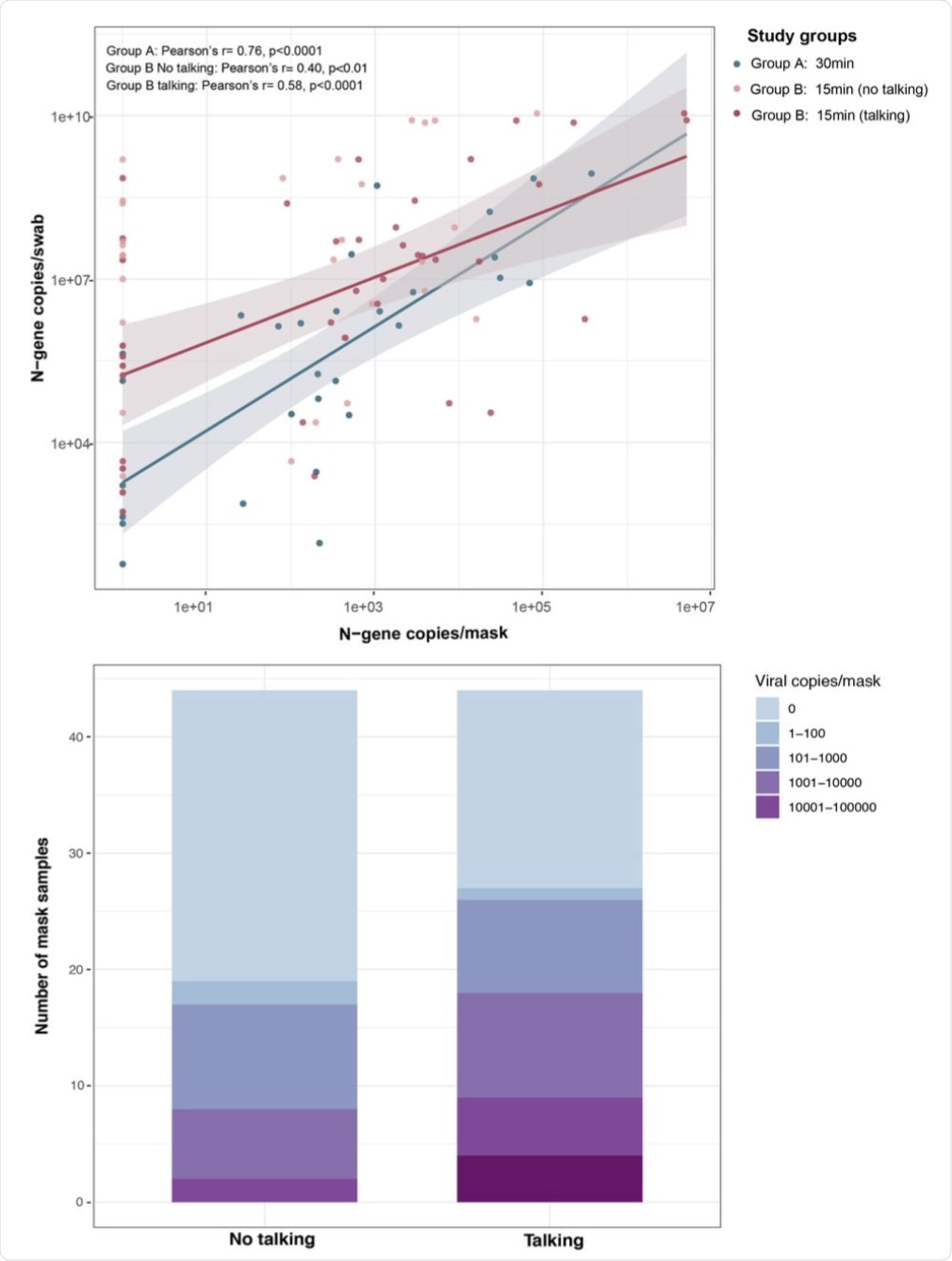 (A) Viral copy numbers detected in masks and nasal swabs were moderately correlated in group A (30 minutes sampling without instructions; blue, n=33) and group B (15 minutes sampling; red, n=44) talking cohort. (B) Total number of viral copies detected per mask sample in talking and no talking group. The viral copies were detected using SARS-CoV-2 specific N1 and N2 probes. Viral copies were quantified on standard curve derived from N-gene containing pET21b+ plasmid.
(A) Viral copy numbers detected in masks and nasal swabs were moderately correlated in group A (30 minutes sampling without instructions; blue, n=33) and group B (15 minutes sampling; red, n=44) talking cohort. (B) Total number of viral copies detected per mask sample in talking and no talking group. The viral copies were detected using SARS-CoV-2 specific N1 and N2 probes. Viral copies were quantified on standard curve derived from N-gene containing pET21b+ plasmid.
By sequencing the exhaled breath samples, the researchers assessed the quality of RNA extracted from the mask filters and determined whether the mask sampling can be used for genomic surveillance. To this end, the researchers found that 71% of all mask samples met their targets for sequencing coverage, thereby demonstrating the potential application of bioaerosol sampling for the whole genome sequencing of respiratory pathogens. This mask sampling tool is therefore a promising method to study transmission and variant detection.
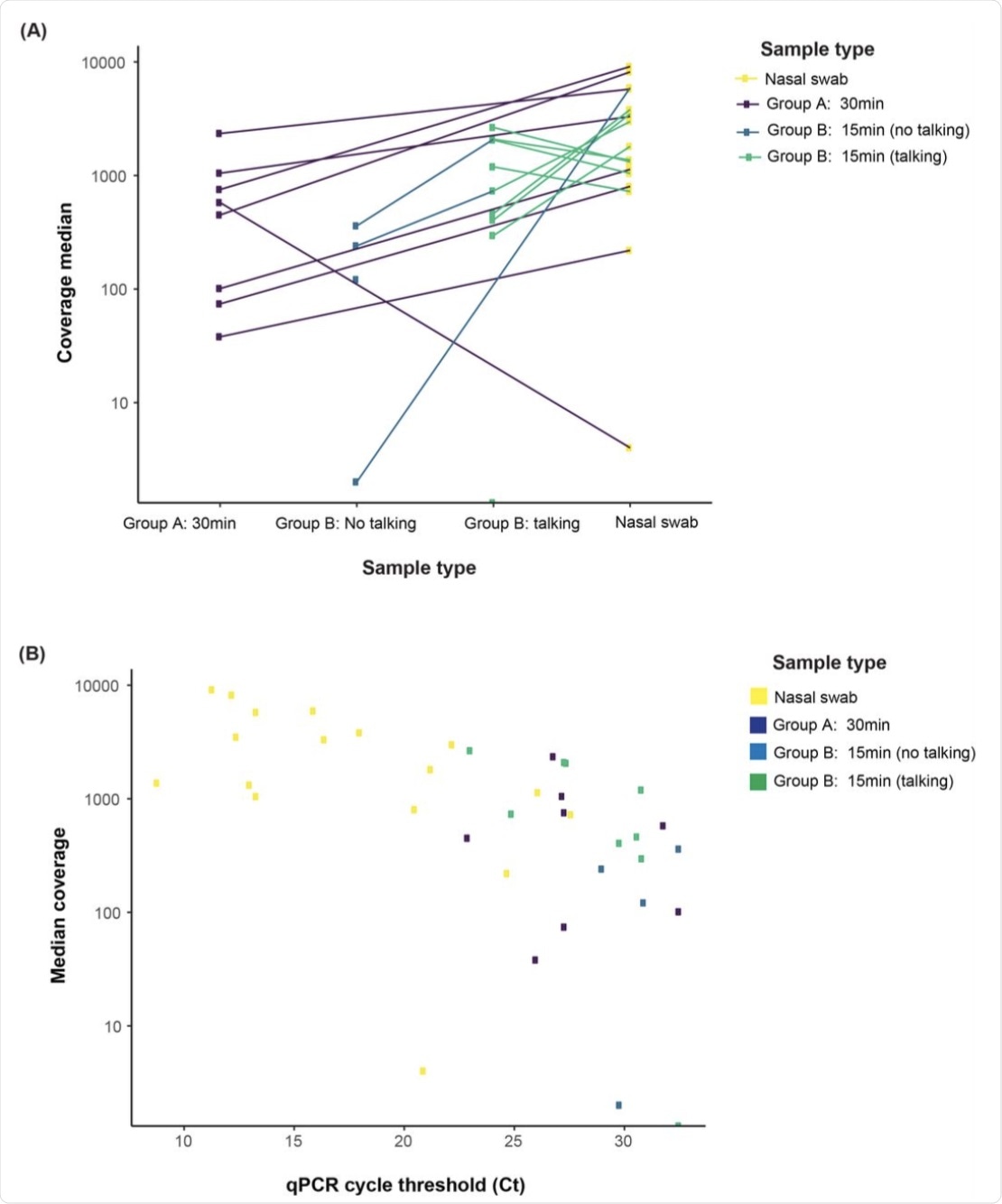 (A) The majority 71.4% (15/21) of all mask samples, including 62.5% (5/8) mask, 50.0% mask no talking, and 88.9% (8/9) mask talking samples, met our targets for sequencing coverage depth (100X) and width (>90% of the genome with >10X coverage). (B) Median coverage depth was correlated with viral load (r = 0.65, p < 0.001).
(A) The majority 71.4% (15/21) of all mask samples, including 62.5% (5/8) mask, 50.0% mask no talking, and 88.9% (8/9) mask talking samples, met our targets for sequencing coverage depth (100X) and width (>90% of the genome with >10X coverage). (B) Median coverage depth was correlated with viral load (r = 0.65, p < 0.001).
Conclusion
In the current study, the researchers developed a novel mask sampling tool that can be used to quantify and sequence SARS-CoV-2 genetic material from exhaled breath samples. The data presented here also provides evidence on the impact of speech and interindividual variation on SARS-CoV-2 shedding in bioaerosols.
The study focused on an unaddressed area of the quantitative assessment of SARS-CoV-2 bioaerosol production and its determinants. It helps to inform assessment and risk stratification of exposures and bioaerosol infection transmission, as well as close contact guidelines. Exposure varies substantially between individuals but might be risk-stratified by nasal swab viral load and whether the exposure involved conversation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Renu, V., Kim, E., Degner, N., et al. (2021). Variation in SARS-CoV-2 bioaerosol production in exhaled breath. medRxiv. doi:10.1101/2021.07/20.21260892. https://www.medrxiv.org/content/10.1101/2021.07.20.21260892v1
- Peer reviewed and published scientific report.
Verma, Renu, Eugene Kim, Nicholas Degner, Katharine S Walter, Upinder Singh, and Jason R Andrews. 2021. “Variation in Severe Acute Respiratory Syndrome Coronavirus 2 Bioaerosol Production in Exhaled Breath.” Open Forum Infectious Diseases 9 (1). https://doi.org/10.1093/ofid/ofab600. https://academic.oup.com/ofid/article/9/1/ofab600/6447624.