The Centers for Disease Control and Prevention (CDC) issued a letter detailing a case of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in an immunocompromised patient who did not respond well to monoclonal antibody treatment. The resistance to treatment stemmed from an escape mutation on the spike protein known as Q493R.
Currently, Q493R/K is the only mutation that causes resistance to bamlanivimab and etesevimab. It also confers resistance against class 3 monoclonal antibodies “that do not overlap with the angiotensin-converting enzyme 2 (ACE2) binding site and have accessibility to the receptor-binding domain epitope in the up and down conformations.”
According to the researchers, genomic surveillance of SARS-CoV-2 variants is recommended for patients who respond poorly to spike protein monoclonal antibodies. More research is also needed to identify other escape mutations that may emerge after treatment.
The research letter is published in the CDC’s journal Emerging Infectious Diseases.
Case study of severe COVID-19 infection in an immunocompromised patient
On April 12, 2021, a 73-year-old with cholangiocarcinoma was admitted to a hospital in Italy after developing sepsis during one of his chemotherapy cycles. Upon admission, a PCR test was administered where he tested negative for SARS-CoV-2. As a result, for his sepsis, the patient received steroid and antimicrobial drugs.
On April 24, the patient tested positive for SARS-CoV-2 on a separate PCR test. A day after recovering from sepsis, the patient moved to the coronavirus wing of the hospital, where he qualified for emergency use of spike protein monoclonal antibodies.
Two days later, the patient was administered a single intravenous infusion consisting of 700 mg of bamlanivimab and 400 mg of etesevimab. Later PCR tests continued to show a positive result for SARS-CoV-2 with a rising viral load.
On April 30, a chest scan revealed the patient developed interstitial pneumonia, and the decision was made to deliver noninvasive ventilation. Monoclonal antibody treatment ceased during this time.
The patient passed away on May 14, 2021.
Genomic sequencing reveals new escape mutation on SARS-CoV-2 spike protein
The team sequenced positive SARS-CoV-2 samples from the deceased patient. Samples from a PCR test on April 24 revealed spike genes related to the Alpha (B.1.1.7) variant. At the time of collection, the Alpha variant comprised 94% of coronavirus infections in Italy.
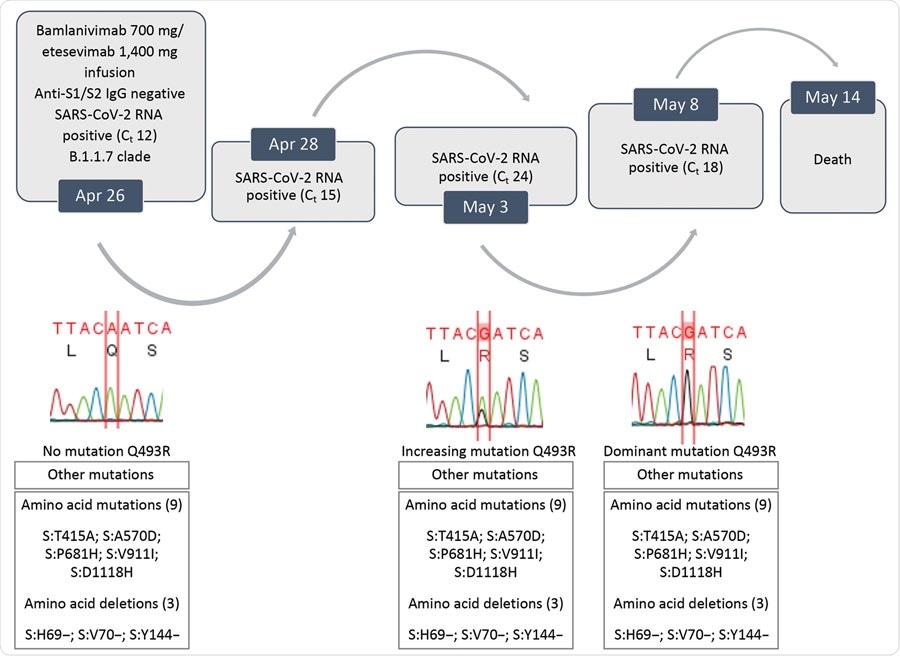
Evolution of SARS-CoV-2 variants in a patient who had coronavirus disease who was given bamlanivimab and etesivimab, Italy. Ct, cycle threshold; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
However, samples collected from a PCR test on May 3 showed a new mutation on the spike protein. It was later revealed to be the Q493R mutation. The Q493R mutation became a predominant mutation in SARS-CoV-2 infections in Italy by May 8.
Further testing revealed the likelihood of getting a coronavirus infection with a Q493R mutation is highly unlikely. The mutation made up 85 out of 1,424,998 genomic sequences as of May 8, 2021, or 0.006%.
Q493R mutation confers resistance to monoclonal antibody treatment
The Q493R mutation makes monoclonal antibody treatment less effective by disrupting its binding to the spike protein.
Bamlanivimab needs E484, F490, Q493, and S494 amino acid residues to bind with for treatment. Q494 is a residue that is also critical for etesevimab binding.
Experiments testing the neutralization power of monoclonal antibodies against this mutation showed Q493R made bamlanivimab less effective by more than 6,666-fold. In addition, Q493R made etesevimab less effective by 232-fold.
Combined monoclonal antibodies were less effective against a pseudovirus with the Q493R mutation by more than 100-fold.