Researchers in the United States have demonstrated the potential of a monoclonal antibody to inform the design of pan-coronavirus vaccines that could prevent the outbreak of future pandemics such as coronavirus disease 2019 (COVID-19).
The team from the Fred Hutchinson Cancer Research Center in Seattle, Washington and The Scripps Research Institute in La Jolla, California, had already shown that the antibody – called CV3-25 –neutralizes the B.1.351 (beta) variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Furthermore, the antibody cross neutralized SARS-CoV-1 and displayed cross-reactive binding to recombinant proteins derived from the human coronaviruses OC43 and HKU.
Now, Andrew McGuire and colleagues have also shown that CV3-25 neutralizes the B.1.1.7 (alpha), B.1.617.2 (delta) and P.1 (gamma) SARS-CoV-2 variants of concern, as well as a SARS-CoV-like bat coronavirus with the potential for zoonotic (animal to human) spread.
The team reports that the antibody binds to a conserved linear peptide within the stem-helix region of sarbecovirus spike proteins. Sarbecovirus refers to the viral subgenus that includes SARS-CoV-1 and SARS-CoV-2, and the spike protein is the main surface structure that coronaviruses use to bind to and infect host cells.
The study found that CV3-25 binds to an epitope within the stem helix that is distinct from those of other monoclonal antibodies that target this region.
“Thus, CV3-25 defines a novel site of sarbecovirus vulnerability that will inform pan-coronavirus vaccine development,” writes McGuire and colleagues.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Three highly pathogenic coronaviruses have emerged in human populations this century
Over the past two decades, three highly pathogenic coronaviruses have arisen in human populations and caused significant morbidity and mortality. The most current and widespread of these is SARS-CoV-2, following the emergence of SARS-CoV-1 in China in 2002 and Middle East Respiratory Syndrome (MERS) in Saudi Arabia in 2012.
“The fact that SARS-CoV-2 is the third highly pathogenic coronavirus to cause significant loss of human life in the past two decades suggests that future coronavirus outbreaks are plausible, if not inevitable,” said the researchers. “Due to extensive coronavirus genetic diversity, the wide range of animal hosts, and potential for zoonotic transmission, there is a need for vaccines and therapeutic agents that can prevent or limit future outbreaks.”
More about the coronavirus infection process
Coronavirus infection is mediated by the viral spike protein, which is comprised of two distinct subunits. The S1 subunit contains the N-terminal domain (NTD) and the receptor-binding domain (RBD) that interacts with the host cell receptor angiotensin-converting enzyme 2 (ACE2). The S2 domain contains the machinery required for the fusion of the host cell and viral membranes.
The neutralizing antibodies elicited by SARS-CoV-2 vaccination or natural infection are an important correlate of protection against subsequent coronavirus infection. The primary targets of these neutralizing antibodies are the RBD and NTD within the S1 subunit.
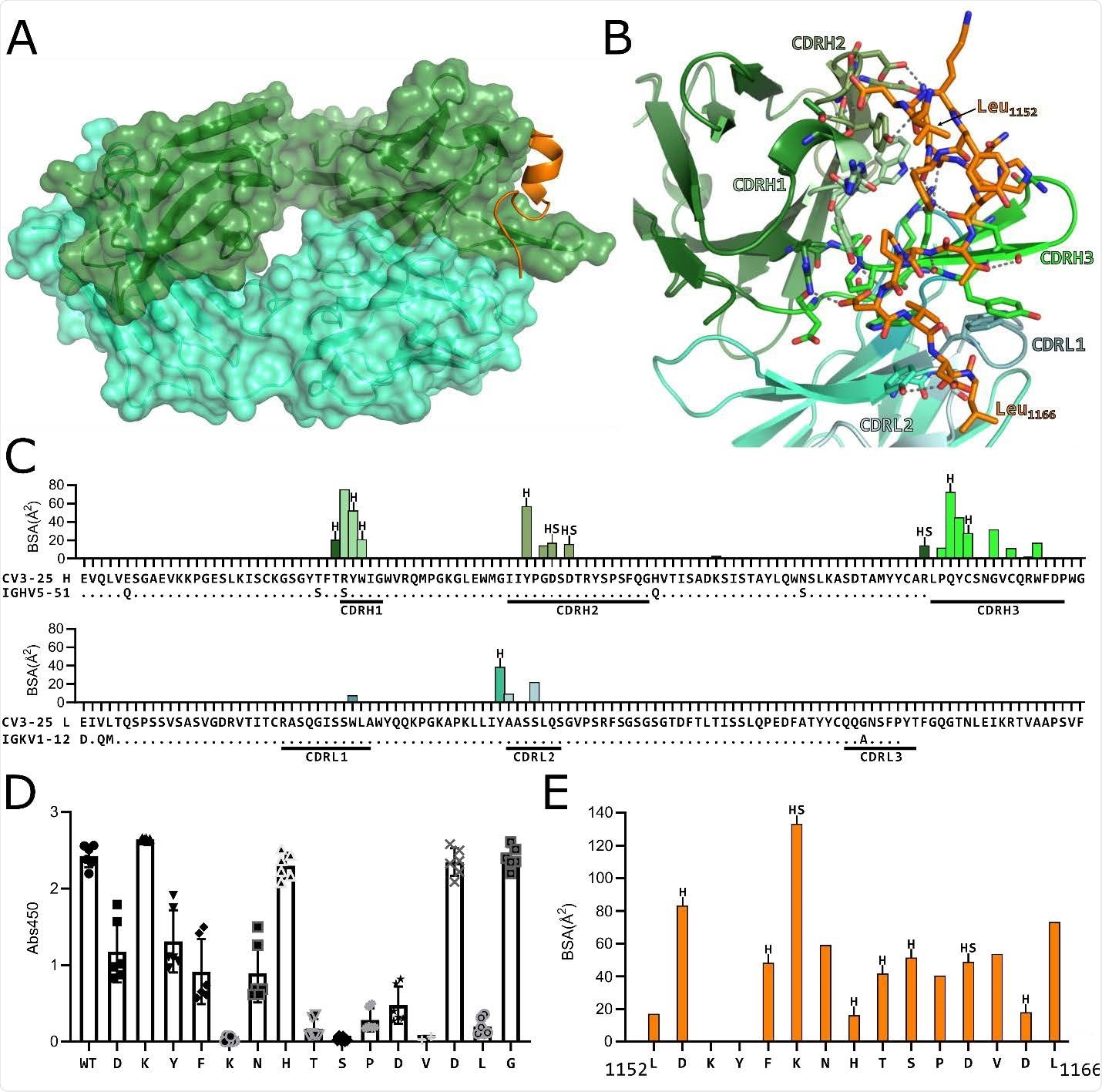
(A) Structure of CV3-25 Fab bound to stem helix peptide. CV3-25-peptide shown in ribbon structure with mAb surface representation shown in transparency. CV3- 25 heavy chain is shown in green and light chain in cyan. The peptide is shown in orange. (B) Details of the interactions between the Fab and the peptide. Complementary determining regions (CDRs) are labeled and colored as shown. Hydrogen bonds between Fab residues and the peptide are shown with dashed black lines. (C) Plots of buried surface area (BSA) of each Fab residue interacting with the peptide and a sequence alignment with the corresponding V-gene. CDRs are labelled and color coded to match the structure shown in B. Residues engaged in a hydrogen bond or salt bridge are marked with an “H” or “S”, respectively. (D) Alanine scanning plot of the stem helix region that CV3-25 binds. CV3-25 binding to linear peptides corresponding to amino acids 1153-1167 of the SARS-CoV-2 spike, where each amino acid was substituted by alanine was measured by ELISA. The absorbance at 450 nm resulting from the addition of 1.25 µg of CV3-25 is shown. Each dot represents a technical replicate from three independent experiments conducted in duplicate. Full titrations are shown in Figure S1. (E) Plot of the BSA of each stem helix peptide residue.
However, due to the variability of spike sequences across coronaviruses, monoclonal antibodies directed at the RBD and NTD are often virus-specific.
“Even within the same coronavirus, mutant variants can evade neutralization by monoclonal antibodies and polyclonal sera,” says McGuire and colleagues. “Indeed, mutations found in the RBD and NTD of SARS-CoV-2 variants of concern are responsible for increased resistance to serum and monoclonal antibodies.”
By contrast, the S2 spike subunit is more functionally and structurally conserved across the different coronaviruses.
“The isolation and characterization of monoclonal antibodies that target conserved neutralizing epitopes across coronavirus variants and strains could inform the design of pan-coronavirus vaccines to help prevent or blunt future outbreaks,” writes the team.
What did the researchers do?
The researchers previously showed that CV3-25 binds to the fusion machinery within S2, neutralizes the B.1.351 variant of concern, cross neutralizes SARS-CoV-1 and shows cross-reactivity with recombinant spike proteins derived from the coronaviruses OC43 and HKU1.
Now, the team has assessed the ability of CV3-25 to neutralize the SARS-CoV-2 variants of concern B.1.1.7, B.1.617.2 and P.1, as well as a more distantly related SARS-like bat coronavirus called WIV1. The latter uses ACE2 as an entry receptor and has been shown to infect human cell lines, thereby representing a bat coronavirus with pandemic potential.
McGuire and colleagues found that CV3-25 neutralized all of the variants and WIV1 with comparable potency.
“Combined with the observation that CV3-25 also neutralizes SARS-CoV-1, these data indicate that it binds to an epitope on S2 that is unaffected by mutations found in these variants,” they write.
A distinct epitope that will inform the design of pan-coronavirus vaccines
Next, the team showed that CV3-25 maintained this neutralizing activity across the different variants by binding to a conserved linear peptide within the stem-helix region on sarbecovirus spike proteins.
A crystal structure of a CV3-25–peptide complex revealed that the antibody binds to a solvent-exposed linear epitope that partially unwinds the stem-helix. This CV3-25 epitope is distinct from those of other stem-helix-directed monoclonal antibodies, say the researchers.
McGuire and colleagues say that CV3-25, therefore, defines a novel site of conserved vulnerability that will enable the development and design of pan-coronavirus vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
McGuire A, et al. Structural definition of a pan-sarbecovirus neutralizing epitope on the spike S2 subunit. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.08.02.454829, https://www.biorxiv.org/content/10.1101/2021.08.02.454829v1
- Peer reviewed and published scientific report.
Hurlburt, Nicholas K., Leah J. Homad, Irika Sinha, Madeleine F. Jennewein, Anna J. MacCamy, Yu-Hsin Wan, Jim Boonyaratanakornkit, et al. 2022. “Structural Definition of a Pan-Sarbecovirus Neutralizing Epitope on the Spike S2 Subunit.” Communications Biology 5 (1): 1–13. https://doi.org/10.1038/s42003-022-03262-7. https://www.nature.com/articles/s42003-022-03262-7.