Researchers in the United States and Canada have described the structural basis and mode of action for two monoclonal antibodies that potently neutralize severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
Walther Mothes from Yale University School of Medicine in New Haven, Connecticut, and colleagues report that the two antibodies – CV3-1 and CV3-25 – were effective against SARS-CoV-2 variants of concern both in vitro and in vivo.
The researchers say the antibodies were, therefore, prime candidates for elucidating mode of action and identifying epitopes with pan-coronavirus activity.
“We believe that both epitopes of these two antibodies are of interest for passive and active immunization strategies against emerging variants,” they write.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The emergence of SARS-CoV-2 variants threatens vaccination efforts
The SARS-CoV-2 virus is the third betacoronavirus to have emerged in the human population since 2003 and is responsible for the COVID-19 pandemic that remains uncontrolled in many countries due to the emergence of novel variants.
The viral spike protein, which mediates the initial stage of the infection process, is a primary target of the host immune response following natural infection. Therefore, this spike immunogen forms the basis of the Moderna, Pfizer-BioNTech, Johnson & Johnson and AstraZeneca vaccines that are currently being rolled out in many countries.
However, the B.1.1.7 (alpha), B.1.351 (beta), P.1 (gamma), and B.1.617.2 (delta) strains all exhibit increased transmissibility and escape from vaccine and infection-induced immunity compared with previously circulating strains.
More about the structure of the spike
The spike is comprised of two subunits. The S1 subunit contains the receptor-binding domain (RBD) that interacts with the host cell receptor angiotensin-converting enzyme 2 (ACE2), while S2 contains the machinery required for host-viral membrane fusion following S1 shedding.
Cryo-electron microscopy (cryoEM) cryo-electron tomography 70 (cryoET) has previously revealed several distinct pre-fusion conformations wherein the RBD adopts “up” or “down” configurations. The ACE2 receptor binds to and stabilizes RBD in the up configuration.
Antibodies isolated from convalescent or vaccinated individuals can be classified by their specificity for three main epitopes: the RBD, the N-terminal domain (NTD), and the S2 subunit. For each antibody class, the conformational preference for either the RBD-up or RBD-down configurations have been described. Antibodies targeting the RBD are often less potent against emerging variants of concern due to the immune escape mutations that have evolved in this domain.
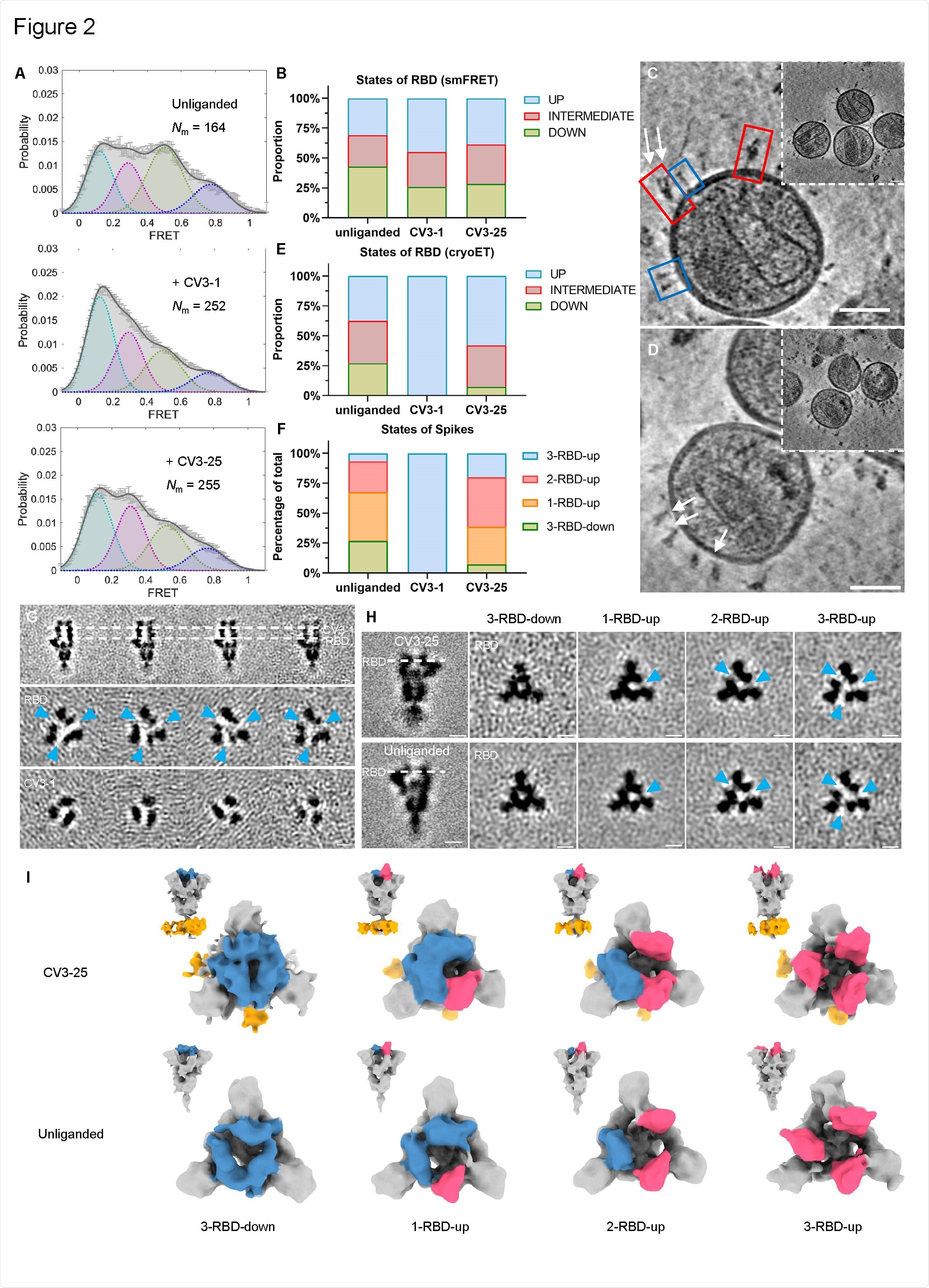
Conformational Dynamics of SB.1.1.7 Bound with CV3-1 and CV3-25. (A) Conformational states of SB.1.1.7 on lentiviral particles monitored by smFRET for unliganded, CV3-1 and CV3-25 bound SB.1.1.7. FRET histograms with number (Nm) of individual dynamic molecules/traces compiled into a conformation-population FRET histogram (gray lines) and fitted into a 4-state Gaussian distribution (solid black) centered at 0.1-FRET (dashed cyan), 0.3- FRET (dashed red), 0.5-FRET (dashed green), and 0.8-FRET (dashed magenta). (B) Proportion of different states of RBD identified by smFRET in (A). For parallel comparative to cryoET data, 0.8-FRET portion was omitted, due to its structural uncertainty. (C-D) Zoomed-in views of SARS-CoV-2 pseudoviruses bearing S bound by CV3-1 (C) and CV3- 25 (D) Fabs and representative slices of tomograms (insets). Scale bar, 50 nm. White arrows indicate bound Fabs. Red boxes, prefusion spikes; Blue boxes, post-fusion spikes. (E) Proportion of different states of RBD at different conditions from cyroET data. UP state was separated by focused classification on the RBD region. The remaining of the RBDs were defined as DOWN state if there was no up RBD on the same spike, otherwise, they were considered as an INTERMEDIATE state. (F) Proportion of different RBD states of spikes on virions with and without Fabs bound. Spikes were grouped into 3-RBD-down,1-RBD-up, 2-RBD-up and 3-RBD-up classes. (G) Side views (top panel) and top views (middle and bottom panels) of subclasses of averaged S bound by CV3-1 Fabs. (H) Side views (left column) of the consensus structure of unliganded (bottom) and CV3-25 bound (top) S and top views of subclass averages (right columns) obtained after focused classification on the RBD of S. In (G-H), dotted lines indicate the positions of top-view sections. Blue arrowheads point to the gap in density between RBD and the neighboring NTD that appears when the RBD moves into the UP-state. Scale bar, 5 nm. (I) Segmentation of subclass averages of unliganded (bottom) and CV3-25 bound (top) S. Top views and side views (insets) are shown for 3-RBD-down, 1-RBD-up, 2-RBD-up and 3-RBD-up classes. Down RBDs and up RBDs are shown in blue and red respectively, CV3-25 Fabs are shown in orange.
The potential for cross-reactive antibodies and vaccines
Although the immune responses induced by the current vaccines provide varying degrees of protection against all known variants of concern, a booster shot may be needed to ensure sufficient protection against future variants.
Furthermore, since SARS-CoV-2 is the third betacoronavirus after SARS-CoV-1 and Middle East Respiratory Syndrome (MERS) to arise in humans this century and given the sizeable natural reservoir of similar viruses in wildlife species such as bats, another coronavirus pandemic is likely to occur in the future.
The S2 domain of these coronaviruses is highly conserved, which raises the possibility of cross-reactive antibodies and vaccines, says Mothe and colleagues.
The researchers previously characterized two potent spike-binding antibodies – CV3-1 and CV3-25 – isolated from convalescent patients. While CV3-1 specifically targets the RBD, CV3-25 recognizes the S2 domains of several betacoronaviruses.
What did the current study involve?
First, the team tested the ability of the antibodies to recognize and neutralize the B.1.1.7, B.1.351, P.1, and B.1.617.2 variants of concern as well as the B.1.429 (epsilon), B.1.525 (eta), B.1.526 (iota) and B.1.617.1 (kappa) variants of interest.
The CV3-1 antibody effectively bound to cells expressing the spike proteins from these different variants and exhibited potent neutralizing activity.
The CV3-25 antibody was less potent but still effectively bound and neutralized all variants.
Both antibodies also limited viral replication in the nose and lungs of K18-hACE2 mice that were delivered a lethal dose of the B.1.351 variant.
“These data demonstrate that in contrast to other antibodies that are attenuated against emerging variants,CV3-1 and CV3-25 remain potent against these variants and are therefore prime candidates to elucidate the mode of action and identify epitopes with pan-coronavirus activity,” writes Mothes and colleagues.
Investigating the antibodies’ structure and mode of action
Next, the researchers elucidated the structure and mode of action for the two antibodies by performing cryoET of virus-like particles (VLPs) carrying the B.1.1.7 variant.
This revealed that CV3-1 bound to the 485-GFN-487 loop of the RBD in the “up” conformation and induced potent shedding of S1.
“The data indicate that CV3-1 is a potent agonist and point to the 485-GFN-487 loop as an allosteric center critical for the activation of S1,” says the team.
By contrast, CV3-25 inhibited membrane fusion by binding to an epitope in the stem helix region of S2.
“Given that the stem helix epitope is highly conserved among β-coronaviruses, immunogens featuring this S2 epitope are interesting candidates for vaccines to cover all variants and possibly exhibit pan-coronavirus efficacy,” writes Mothes and colleagues.
What did the authors conclude?
The researchers say that that the epitopes of both antibodies are of interest for passive and active immunization strategies against emerging variants.
“Vaccine immunogen designs that incorporate the conserved regions in the RBD and stem helix are candidates to elicit pan-coronavirus protective immune responses,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Mothes W, et al. Structural Basis and Mode of Action for Two Broadly Neutralizing Antibodies Against SARS-CoV-2 Emerging Variants of Concern. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.08.02.454546, https://www.biorxiv.org/content/10.1101/2021.08.02.454546v1
- Peer reviewed and published scientific report.
Li, Wenwei, Yaozong Chen, Jérémie Prévost, Irfan Ullah, Maolin Lu, Shang Yu Gong, Alexandra Tauzin, et al. 2022. “Structural Basis and Mode of Action for Two Broadly Neutralizing Antibodies against SARS-CoV-2 Emerging Variants of Concern.” Cell Reports 38 (2). https://doi.org/10.1016/j.celrep.2021.110210. https://www.cell.com/cell-reports/fulltext/S2211-1247(21)01714-9.