Viruses exploit host cell proteins to enter the cell and then propagate, the specific proteins dependent on the virus. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), for example, is known to interact with the angiotensin-converting enzyme 2 (ACE2) receptor and TMPRSS2 protease on the surface of cells to adhere and subsequently undergo cleavage of the spike protein to facilitate cell entry, respectively. Similarly, influenza A must be prepared for receptor binding and fusion by cleavage of the virus' surface hemagglutinin by trypsin-like proteases.
Identifying human proteins required by several diverse virus types would be advantageous in medicine, directing drug discovery efforts towards creating broad-spectrum antivirals.
In a research paper recently uploaded to the bioRxiv* preprint server by Poston et al. (August 8th, 2021), common host factors amongst several zoonotic pathogens are explored by CRISPR-Cas9 screening in human lung cells, focusing in particular on the betacoronavirus genus.
How was the study performed?
SARS-CoV, MERS-CoV, and SARS-CoV-2 are the most prevalent human beta coronaviruses in terms of morbidity. However, several other coronaviruses can infect humans: HCoV-OC43 and HCoV-HKU1 of the betacoronavirus genus, and HCoV-229E and HCoV-NL63 of the alphacoronavirus genus.
The research group began by infecting human lung cells with HCoV-OC43 and incubated them for one week, at which point the surviving cells were assessed for dependency factors. Multiple genes involved in heparan sulfate biosynthesis were identified, suggesting that heparan sulfate is involved in the attachment of the virus to the cell surface, as is known from the literature. In addition, several genes involved endosomal activity and the transport of extracellular material into the cell, including vacuolar protein sorting-associated protein 29 (VPS29), were also identified.
The Rockefeller University research group employed CRISPR-Cas9 gene editing to generate cell lines lacking the identified genes. In these cell lines, cell viability and proliferation were seen to be unaffected, and the cells were subsequently infected with HCoV-OC43, HCoV-NL63, HCoV-229E, or a SARS-CoV-2 pseudovirus, along with several viruses' outside of the coronavirus genus such as respiratory syncytial virus, adenovirus, or influenza A virus.
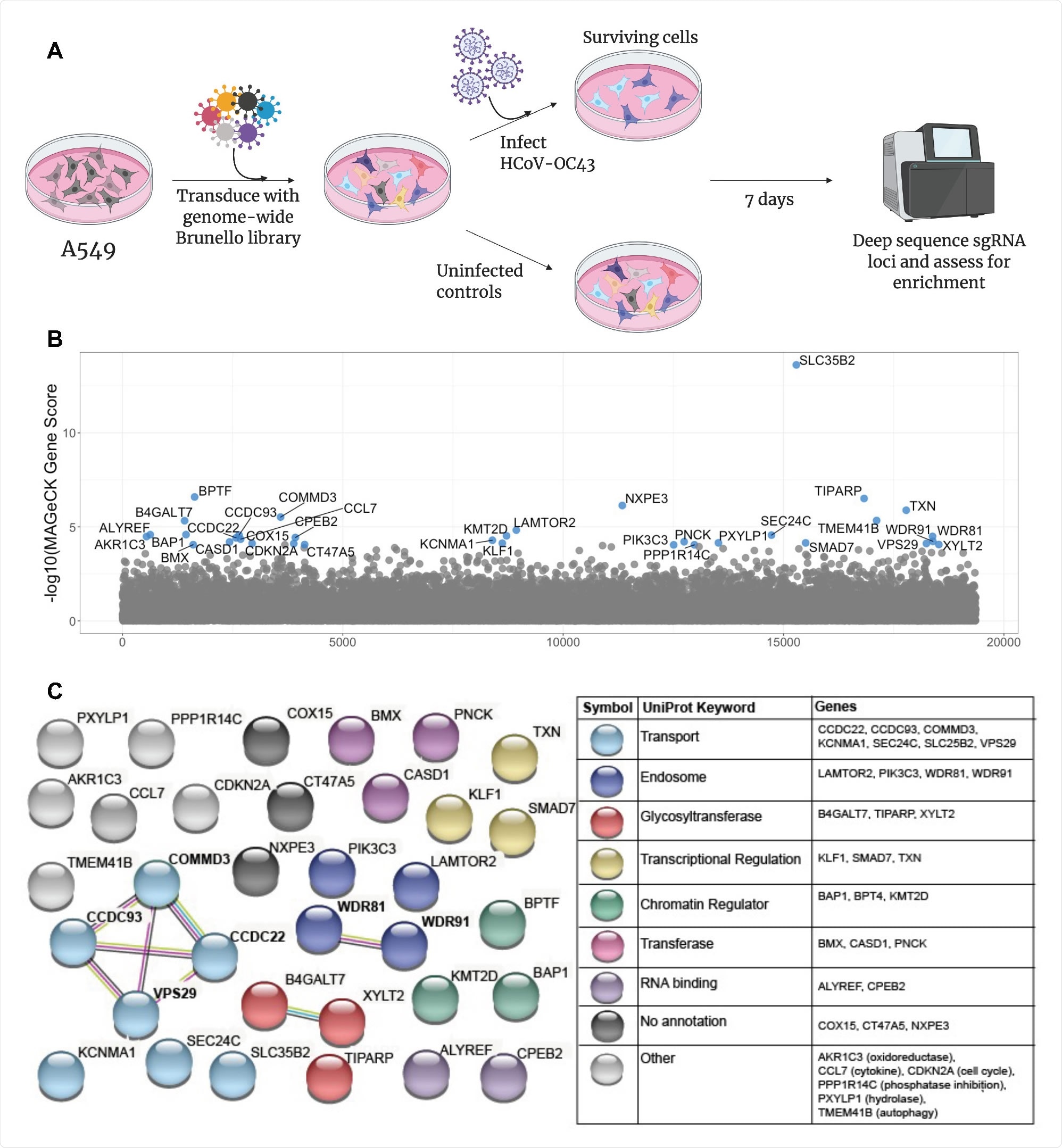
A CRISPR screen reveals genes influencing HCoV-OC43 susceptibility (A) Schematic of screening setup (B) Screen results, where the x-axis corresponds to each unique gene in the library (labeled randomly from 1 to 19,114) and the y-axis denotes the -log10 MAGeCK gene score. All genes scoring higher than the best-scoring non-targeting control pseudogene are labeled in blue. The screen was performed in three independent replicates (C) string-db analysis and UniProt annotation of gene hits. Sphere colors correspond to UniProt keywords and connecting lines indicate strength of evidence underlying gene-gene interactions (pink: experimentally-determined interaction; blue: annotated interaction in curated databases; gray: evidence of co-expression; yellow: text-mining).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
VPS29 knockout
The researchers noted that VPS29 and the associated endosomal regulator complex (CCC complex) were essential in facilitating coronavirus infection across all cell lines tested.
At the same time, the other types of the virus were not impaired by VPS29 knockout. However, as each of the viruses explored is known to exploit endocytic pathways to gain cell entry, this result implies that VPS29 knockout does not generally impair endocytic function within the cell.
Still, that coronavirus entry only is specifically affected. Indeed, human bronchial epithelial primary lung cells with knocked out VPS29 exhibited strong resistance to infection by HCoV-OC43.
In contrast, VPS29 knockout enhanced influenza A virus infection, particularly in two cell lines expressing coronavirus receptors. VPS29 can participate in several protein complexes involved in endocytosis that may preferentially interact with one virus over another. Thus, to investigate this, the group performed a siRNA screen that targeted proteins known to interact with VPS29.
VPS29 can belong to the retromer complex, a key component of endosomal sorting machinery, composed of proteins VPS26A, VPS29, VPS35, or RAB7A. Knockdown of any of these proteins also impaired coronavirus infection, and thus the retromer complex is likely involved in cell entry by coronaviruses. Conversely, influenza A virus infection was observed to be enhanced in the absence of a functioning retromer complex.
The group utilized fluorescent microscopy to determine that VPS29 knockout caused the endosomes to become enlarged and deacidified, though still exhibited normal cargo loading capabilities. A tagged SARS-CoV-2 pseudovirus was used to visualize the route of cell entry, noting that the virus accumulated in endosomes before egress into the cytoplasm in non-knockout cells. While in VSP29 knockout cells, the virus accumulated but was unable to escape the endosome.
Identical experiments with the influenza A virus revealed no accumulation within the endosomes, with egress being enhanced. The group suggests that the higher pH observed within the endosomes of knockout cells is likely the cause of the impaired or enhanced egress into the cytoplasm by coronaviruses or influenza A virus, respectively.
In coronaviruses, several biochemical processing steps must take place to prepare for egress and proliferation, which the change in pH may have disrupted. At the same time, the less acidic environment is simultaneously less destructive towards influenza A virions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources