The entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) into host cells is mediated by interactions between the spike protein of the virus and the angiotensin-converting enzyme 2 (ACE2) cell surface receptor. The spike protein is composed of two subunits. One bears the receptor binding domain (RBD) responsible for interaction with ACE2, while the second bears membrane fusion machinery vital to crossing the cell membrane. The two subunits are connected by a hinge-like furin cleavage site that allows the spike protein to be in an “open” or “closed” conformation, exposing or protecting the RBD, respectively.
The spike protein is also densely coated with glycans that shield the virus from antibodies and play a role in beneficial interactions with other biomolecules, facilitating cell entry and viral propagation. The current conformation of the spike protein, in the open or closed state, also influences the conformation of the glycan coating, and prior computational studies have found that glycans are involved in modulating and stabilizing these transitions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In a paper recently uploaded to the bioRxiv* preprint server, the role of glycan coating on the SARS-CoV-2 spike protein is further investigated by molecular dynamics simulations of the glycosylated and non-glycosylated protein, identifying several key glycans that play a role in stabilizing and protecting the RBD, and also encourage bonding with the ACE2 receptor.
Glycans impart stability
The open and closed states of the spike protein were modeled based on cryo-electron microscopy (cryo-EM) structures and the binding free energy calculated using a replica exchange umbrella sampling approach, wherein copies of the expected glycan are iteratively simulated in differing conformations at each site until a “best fit” is achieved. This process was seeded by mass spectrometry data to identify each glycan, and metadynamic simulations that determine the available space for glycans on the protein surface in either state.
As determined by molecular modeling, the structure of the spike protein at energy minima in the open or closed conformation matched that observed in cryo-EM studies, and the proportion of spike proteins in either conformation was approximately even. When the spike protein is glycosylated, the energy difference between the minima was noted to be small, with the open state just 0.4 kcal/mol higher, explaining the almost equal proportion of conformers observed in simulations. The non-glycosylated spike protein, however, had a lower free energy in the open state by 2.2 kcal/mol, causing a significant shift in the proportion of spike proteins in either state, the population becoming dominated by the open form.
The kinetics of spike protein opening and closing were also assessed by the group in the presence or absence of glycans, with closed-open and open-closed transitions taking 10.1 and 2.5 ms for the glycosylated form, respectively. In contrast, the non-glycosylated protein took only 5 and 305 µs for the same transitions, quicker due to the lower energy barrier. Binding and unbinding rates with the ACE2 receptor were also assessed, finding that at equilibrium, 40% of the glycosylated form are bound with the ACE2 receptor, while the remaining 12% and 48% are in the open state but not bound, or in the closed state, respectively. Non-glycosylated spike protein bound with the ACE2 receptor at even higher rates at equilibrium due to the strong preference for the open conformation, with 77% being bound and the remaining 23% in the open conformation but not bound.
Glycans encourage bonding with ACE2
The N343 glycan of the spike protein has previously been suggested to be essential in the binding of the spike protein with the ACE2 receptor, with mutations to this glycan being detrimental to the binding energy. The group confirms this by simulation, and state that though the overall population of spike protein in the open state is greater following removal of all glycans, the binding affinity towards the ACE2 receptor is actually lower.
The group also notes that a larger number of hydrogen bonds are detected on the glycosylated spike protein in the open conformation than closed. However, in the absence of glycans, an even greater number of hydrogen bonds are present. This can be explained by the interaction of the RBD-bearing spike protein subunit with the opposing subunit, which align at the mid-point between states. When fully open, the hydrogen bonds between these subunits are disrupted by glycans at N165 and N122, which is the reason for the observed strong preference for the open state on the non-glycosylated spike protein.
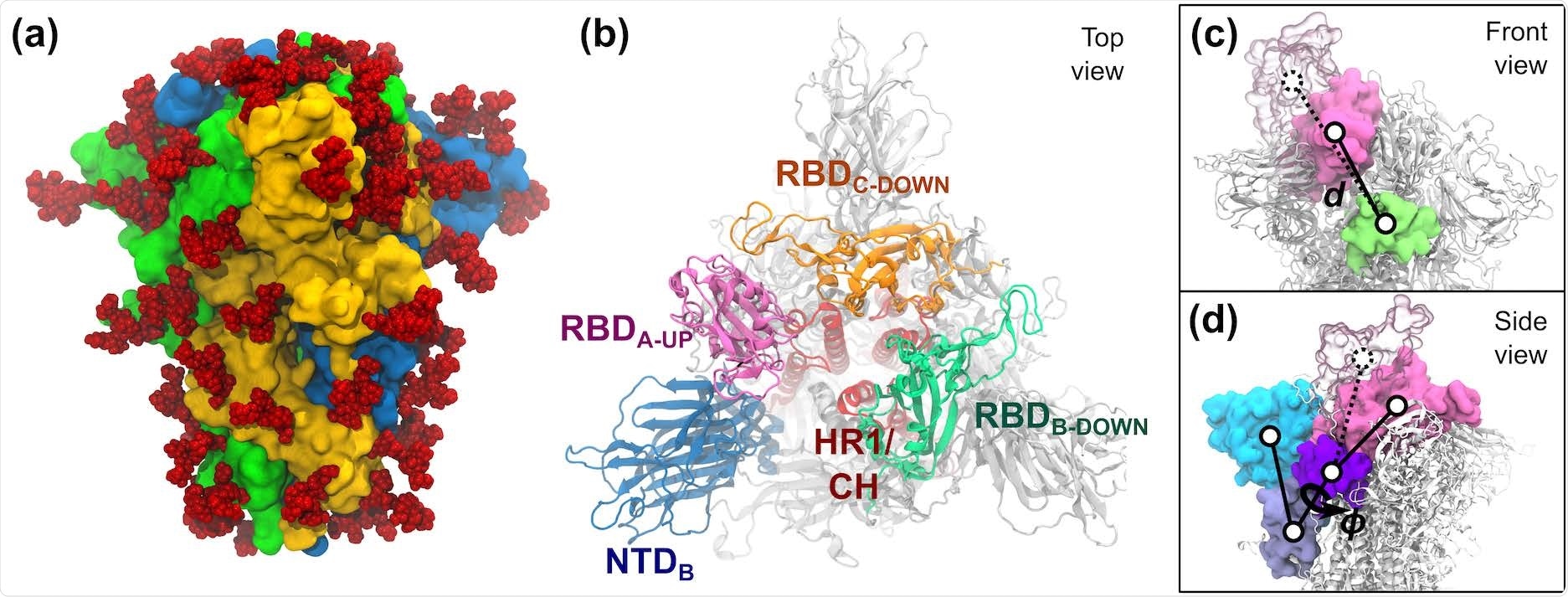
S-protein of SARS-CoV-2. (a) The trimeric S-protein in the all-down state, colored by protomer. Glycans are shown as red spheres. (b) Top view of the S-protein in the one-up state. Important domains of the spike are highlighted, including the N-terminal domain (NTD, 14–306), the receptor binding domains (RBD, 336–518), the heptad repeat 1 (HR1, 908–986), and the central helix (CH, 987–1035). (c,d) The two collective variables defined to describe the opening of RBD-A include: (c) the center-of-mass distance d between RBDA (pink) and SD1-B (lime), and (d) the dihedral angle ϕ formed by the domains RBD-A (pink), SD1-A (purple), SD2-A (ice blue), and NTD-A (cyan). RBD-A in both the down (solid pink) and up (transparent pink) states are shown.
Additional distinct glycan contacts that stabilize the open or closed conformation were noted by the group, including glycans N234 and N343 that changing position notably during the transition from closed to open. Glycan N234 points outward, away from the body of the protein, when in the closed conformation, shifting to an inward-facing conformation as the spike opens. The glycan at N343 also acts as a gate to the RBD, working with the aforementioned glycan N165 to each individually stabilize the closed conformation.
However, glycans N165 and N343 interact significantly when the spike protein is in the open conformation, and work to stabilize it. The group suggest that this interaction also prevents the RBD from prematurely interacting with the adjacent receptor binding motif, which would subsequently inhibit interaction with the ACE2 receptor.
The spike protein, and receptor binding domain, in particular, are also a major target of neutralizing antibodies. However, the RBD is generally inaccessible to antibodies when in the closed conformation due to glycan coverage. The group simulated many known antibodies and examined the binding mode in each conformation, confirming the higher rate of neutralization towards open spike proteins. Interestingly, particular antibodies were identified as having an equally high affinity towards the spike protein when in either conformation, binding to some of the few exposed regions of the RBD, and the group highlights these antibodies for further investigation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources