The coronavirus disease 2019 (COVID-19) pandemic has exposed the persistent threat posed by respiratory viruses. As is evident from the current and previous pandemics, respiratory viruses have an extensive host range and a high risk of animals-to-human spillover.
Interaction — the ability of one pathogen to impact infection or disease caused by another pathogen —has not been studied sufficiently in respiratory viruses. These interactions can be either positive (synergistic or facilitatory) or negative (antagonistic or competitive); for example, up-regulation of viral target receptors or cell fusion are positive interactions and blocking of viral replication caused by the interferon response is a negative interaction.
Different respiratory viruses may have diagonally different effects on COVID-19. While rhinoviruses inhibit severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection via the interferon response, influenza A viruses can facilitate it using upregulation of angiotensin-converting enzyme 2 (ACE2), which is the receptor of SARS-CoV-2 in host cells. SARS-CoV-2 interactions may have vast implications in predicting the future course of the pandemic and also the indirect impacts of non-COVID-19 vaccines on COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Assessing the reliability of epidemiological studies based on co-detection prevalence data in the estimation of interactions among respiratory viruses
Researchers from Germany and France tried to determine if epidemiological studies based on co-detection prevalence data can offer a reliable estimation of interactions among respiratory viruses. They designed a simulation study by building on a general epidemiological model of co-circulation of 2 respiratory viruses that cause seasonal epidemics. This study is currently available on the medRxiv* preprint server while awaiting peer review.
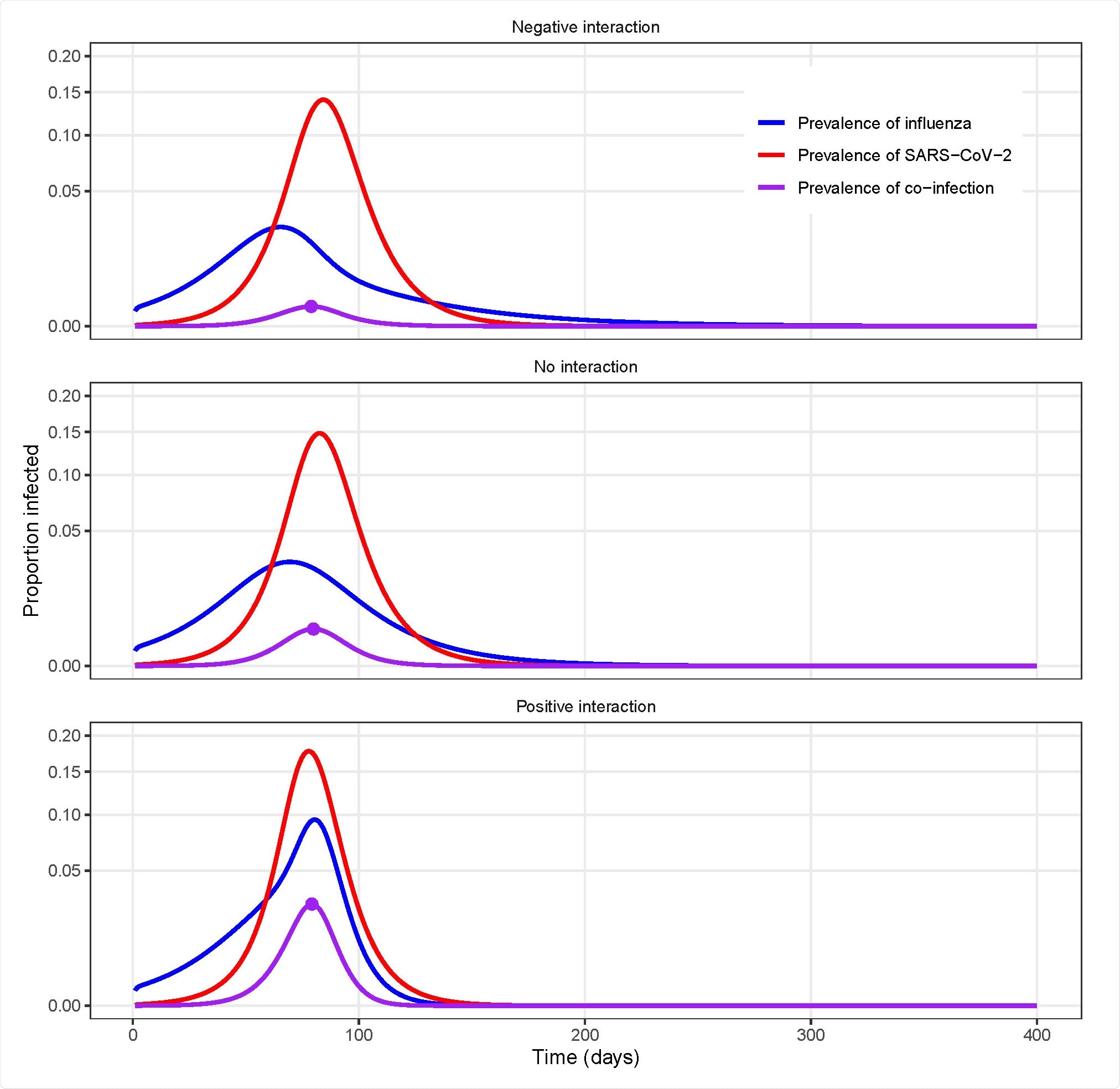
Example model simulations for different types of uniform interactions between influenza and SARS-CoV-2.
The researchers focused on influenza–SARS-CoV-2 pair. They demonstrated that the prevalence ratio systematically underestimates the strength of interaction between the viruses and can even misclassify synergistic or antagonistic interactions that persist after infection clearance. They showed that cross-sectional estimates of co-infection prevalence —either alone or in combination with single-infection prevalence estimates — offer a poor guide to determine interaction. With the help of a global sensitivity analysis, they also identified properties of viral infection, including short infectious period or high reproduction number, that can blur the interaction suggested by the prevalence ratio.
“Our results show that, in the absence of precise information about the timing of interaction, epidemiological studies designed to estimate the prevalence ratio, or variations thereof, may be unreliable.”
Results show that epidemiological studies based on co-detection prevalence data offer a poor guide to interactions between respiratory viruses
Overall, the study results suggest that epidemiological studies based on co-detection prevalence data offer a poor guide to interactions between respiratory viruses. Thus, the authors argue that earlier epidemiological studies based on this design need to be interpreted with caution and more longitudinal studies are needed to explain the epidemiological interactions between SARS-CoV-2 and other respiratory viruses.
“With the likely prospect of COVID-19 becoming endemic, there is a pressing need to elucidate the potential interactions of SARS-CoV-2 with other pathogens, in particular respiratory viruses.”
In conclusion, the findings show that the complex and non-linear dynamic of respiratory viruses makes the interpretation of intuitive interaction measures very difficult. Despite these drawbacks, other statistical or mathematical methods based on longitudinal data should facilitate epidemiological research on viral interactions. According to the authors, with increasing evidence showing SARS-CoV-2 and other pathogens circulate within polymicrobial systems and not in isolation, such research should be prioritized.
“In sum, we submit that further epidemiological studies will be needed to elucidate the interactions of SARS-CoV-2 with other respiratory viruses.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
The pitfalls of inferring virus-virus interactions from co-detection prevalence data: Application to influenza and SARS-CoV-2 Matthieu Domenech de Cellès, Elizabeth Goult, Jean-Sébastien Casalegno, Sarah Kramer, medRxiv, 2021.09.02.21263018; doi: https://doi.org/10.1101/2021.09.02.21263018, https://www.medrxiv.org/content/10.1101/2021.09.02.21263018v1
- Peer reviewed and published scientific report.
Domenech de Cellès, Matthieu, Elizabeth Goult, Jean-Sebastien Casalegno, and Sarah C. Kramer. 2022. “The Pitfalls of Inferring Virus–Virus Interactions from Co-Detection Prevalence Data: Application to Influenza and SARS-CoV-2.” Proceedings of the Royal Society B: Biological Sciences 289 (1966). https://doi.org/10.1098/rspb.2021.2358. https://royalsocietypublishing.org/doi/10.1098/rspb.2021.2358.